diff --git a/docs/images/tutorials/toppview/apply-topp-tool-to-layer.png b/docs/images/tutorials/toppview/apply-topp-tool-to-layer.png
index 0824362e..7b1c797a 100644
Binary files a/docs/images/tutorials/toppview/apply-topp-tool-to-layer.png and b/docs/images/tutorials/toppview/apply-topp-tool-to-layer.png differ
diff --git a/docs/images/tutorials/toppview/data-import-options.png b/docs/images/tutorials/toppview/data-import-options.png
new file mode 100644
index 00000000..55983c0b
Binary files /dev/null and b/docs/images/tutorials/toppview/data-import-options.png differ
diff --git a/docs/images/tutorials/toppview/file-import-options.png b/docs/images/tutorials/toppview/file-import-options.png
new file mode 100644
index 00000000..12cb22e0
Binary files /dev/null and b/docs/images/tutorials/toppview/file-import-options.png differ
diff --git a/docs/images/tutorials/toppview/filtering-options.png b/docs/images/tutorials/toppview/filtering-options.png
new file mode 100644
index 00000000..4ccd8d64
Binary files /dev/null and b/docs/images/tutorials/toppview/filtering-options.png differ
diff --git a/docs/images/tutorials/toppview/layer-loaded-in-viewer.png b/docs/images/tutorials/toppview/layer-loaded-in-viewer.png
new file mode 100644
index 00000000..61e88ac0
Binary files /dev/null and b/docs/images/tutorials/toppview/layer-loaded-in-viewer.png differ
diff --git a/docs/images/tutorials/toppview/layers-window.png b/docs/images/tutorials/toppview/layers-window.png
new file mode 100644
index 00000000..2b0568d2
Binary files /dev/null and b/docs/images/tutorials/toppview/layers-window.png differ
diff --git a/docs/images/tutorials/toppview/select-data-filters-window.png b/docs/images/tutorials/toppview/select-data-filters-window.png
new file mode 100644
index 00000000..e29d32c9
Binary files /dev/null and b/docs/images/tutorials/toppview/select-data-filters-window.png differ
diff --git a/docs/images/tutorials/toppview/topp-tool-arguments-and-description.png b/docs/images/tutorials/toppview/topp-tool-arguments-and-description.png
new file mode 100644
index 00000000..6b763eb4
Binary files /dev/null and b/docs/images/tutorials/toppview/topp-tool-arguments-and-description.png differ
diff --git a/docs/images/tutorials/toppview/topp-tool-parameters.png b/docs/images/tutorials/toppview/topp-tool-parameters.png
new file mode 100644
index 00000000..8ad6e97f
Binary files /dev/null and b/docs/images/tutorials/toppview/topp-tool-parameters.png differ
diff --git a/docs/images/tutorials/toppview/user-interface.png b/docs/images/tutorials/toppview/user-interface.png
new file mode 100644
index 00000000..8e1adf16
Binary files /dev/null and b/docs/images/tutorials/toppview/user-interface.png differ
diff --git a/docs/openms-applications-and-tools/openms-applications.md b/docs/openms-applications-and-tools/openms-applications.md
index 448a6733..2fc7c3b3 100644
--- a/docs/openms-applications-and-tools/openms-applications.md
+++ b/docs/openms-applications-and-tools/openms-applications.md
@@ -1,21 +1,26 @@
OpenMS Applications
===================
-OpenMS provides a suite of applications, designed for users who want easy access to TOPP tools. These applications include:
+OpenMS provides a suite of graphical applications, designed for users who want easy access to TOPP tools. These applications include:
+
- **INIFileEditor**
- A GUI application used to edit INI files. INI files are files with the extension .ini and are common configuration files for all TOPP tools.
-
-- **TOPP shell**
+ A GUI application used to edit INI files. INI files are used to configure TOPP tool parameters. INI files are files with the extension `.ini`. For more information, read our [INIFile Editor](openms-applications/ini-file-editor.md) section.
- A Command Line Interface (CLI) that provides easy access to tools. Users don’t need to use TOPP shell; instead, they can configure their own CLI to directly use TOPP tools.
- **TOPPView**
- A GUI application used to inspect, visualize and compare mass spectrometry data. Read more in-depth documentation about TOPPView.
+ A GUI application used to inspect, visualize and compare mass spectrometry data. Read more in-depth documentation about TOPPView. For more information, read our [TOPPView: Visualize with OpenMS](visualize-with-openms.md) section.
- **TOPPAS (deprecated)**
A GUI application used to apply multiple tools sequentially on mass spectrometry data. Applying multiple tools in a sequence is referred to as a workflow or a pipeline. OpenMS no longer supports TOPPAS and instead recommends the use of [KNIME](https://www.knime.com/), for which we provide a community plugin.
- **SwathWizard**
-An application for SWATH analysis. SwathWizard is used to analyze DIA swath data.
+ An application for SWATH analysis. SwathWizard is used to analyze DIA swath data. For mor information, read our [SwathWizard](openms-applications/swathwizard.md)
+
+
+A typical workflow would consist of the following steps:
+
+1. Edit the INI file in the INIFile Editor.
+2. Import data into TOPPView.
+3. Apply TOPP tool to data in TOPPView. You will need to load the INI file edited in step 1.
diff --git a/docs/graphical-topp-tools/ini-file-editor.md b/docs/openms-applications-and-tools/openms-applications/ini-file-editor.md
similarity index 57%
rename from docs/graphical-topp-tools/ini-file-editor.md
rename to docs/openms-applications-and-tools/openms-applications/ini-file-editor.md
index 541316c1..326225a1 100644
--- a/docs/graphical-topp-tools/ini-file-editor.md
+++ b/docs/openms-applications-and-tools/openms-applications/ini-file-editor.md
@@ -1,10 +1,10 @@
INIFileEditor
============
-Can be used to visually edit INI files of TOPP tools.
+The INIFileEditor is an application used to visually edit INI files of TOPP tools.
The values can be edited by double-clicking or pressing F2.
The documentation of each value is shown in the text area on the bottom of the widget.
-
+
diff --git a/docs/graphical-topp-tools/swathwizard.md b/docs/openms-applications-and-tools/openms-applications/swathwizard.md
similarity index 93%
rename from docs/graphical-topp-tools/swathwizard.md
rename to docs/openms-applications-and-tools/openms-applications/swathwizard.md
index 22412fdd..962f4f76 100644
--- a/docs/graphical-topp-tools/swathwizard.md
+++ b/docs/openms-applications-and-tools/openms-applications/swathwizard.md
@@ -1,7 +1,7 @@
SwathWizard
-==========
+============
-An assistant for Swath analysis.
+SwathWizard is an assistant for Swath analysis.
The Wizard takes the user through the whole analysis pipeline for SWATH proteomics data analysis, i.e. the
[TOPP Documentation: OpenSwathWorkflow](https://abibuilder.informatik.uni-tuebingen.de/archive/openms/Documentation/nightly/html/UTILS_OpenSwathWorkflow.html) tool, including downstream tools such as [GitHub:PyProphet/pyProphet](https://github.com/PyProphet/pyprophet) and the [GitHub:msproteomicstools/TRIC alignment](https://github.com/msproteomicstools/msproteomicstools) tool.
@@ -16,7 +16,7 @@ both the intermediate files from OpenSWATH (e.g. the XIC data in `.sqMass` forma
This is how the wizard looks like:
-
+
A schematic of the internal data flow (all tools are called by SwathWizard in the background) can be found in the
[TOPP Documentation: SwathWizard](https://abibuilder.informatik.uni-tuebingen.de/archive/openms/Documentation/nightly/html/TOPP_SwathWizard.html).
diff --git a/docs/openms-applications-and-tools/topp-tools.md b/docs/openms-applications-and-tools/topp-tools.md
index 49596b8d..a4d3d2fb 100644
--- a/docs/openms-applications-and-tools/topp-tools.md
+++ b/docs/openms-applications-and-tools/topp-tools.md
@@ -7,7 +7,6 @@ The following sections introduce TOPP, some general concepts and more specific i
:maxdepth: 1
topp-tools/introduction-to-topp.md
-topp-tools/key-concepts.md
topp-tools/types-of-topp-tools.md
```
diff --git a/docs/openms-applications-and-tools/topp-tools/introduction-to-topp.md b/docs/openms-applications-and-tools/topp-tools/introduction-to-topp.md
index c4217c7b..7942ba09 100644
--- a/docs/openms-applications-and-tools/topp-tools/introduction-to-topp.md
+++ b/docs/openms-applications-and-tools/topp-tools/introduction-to-topp.md
@@ -1,6 +1,66 @@
Introduction to TOPP
====================
-OpenMS provides a vast array of tools called TOPP tools that automate typical mass processing tasks. They can be either:
-- Executed from the command line or,
-- Applied using graphical applications.
+**TOPP - The OpenMS Pipeline** is a set of tools for the analysis of HPLC-MS data. These tools can be either:
+
+- [Executed from the command line](../openms-applications-and-tools/command-line-interface.md) or,
+- Applied individually using OpenMS graphical applications.
+- Applied in sequence as a workflow using a workflow editor such as KNIME, Nextflow or Galaxy.
+
+Before you choose one of the above options, there are few concepts that need to be understood.
+
+## File formats
+
+OpenMS only accepts files in the following formats:
+
+- **mzML**: The HUPO-PSI standard format for mass spectrometry data.
+- **featureXML**: The OpenMS format for quantitation results.
+- **consensusXML**: The OpenMS format for grouping features in one map or across several maps.
+- **idXML**: The OpenMS format for protein and peptide identification.
+
+Documented schemas of the OpenMS formats can be found [here](https://github.com/OpenMS/OpenMS/tree/develop/share/OpenMS/SCHEMAS).
+
+If your data is not in the above formats, you may need to use one of a file conversion TOPP tool.
+
+## TOPP INI files
+
+TOPP INI files are XML-based files with an `.ini` extension. OpenMS uses TOPP INI files to set parameters for one or more TOPP tools. Alternatively, the command line can be used to set TOPP tool parameters.
+Here is an example of a TOPP INI file:
+
+ ```xml
+
+
+
+
+
+ - Loading an INI file by clicking Load and selecting an INI File from the file importer.
+
+ - Editing the parameters shown in the table and then saving the INI file. To edit a parameter, double click a row in the table and enter a value or choose from the options available. The modified value will be highlighted yellow. To save the parameters, click Store and enter a file name for the INI file.
+
+
+ 
+5. Click **Ok**. You will be prompted to load the new dataset as a **new window** or a **new layer**. Choose an option and click **Ok**.
+
+ 
+
+6. If you chose to load the data in a new window, a new tab will appear. To view that data, select the tab. If you chose to load the data as a new layer, the data will be visualized in the **Viewer**. You can also see the new layer wihout a name in the **Layers window**.
+
+ 
+
+7. (Optional) If you did choose to import the data as a new layer, give the new layer a name. To do this, right-click the layer in the **Layers window** and select **Rename**. Enter a name and click OK.
+
+### Filter data
+
+You may only want to see some data from your dataset and hide the rest. OpenMS allows you to filter data based on the following fields: **Intensity**, **Quality**, **Charge**, **Size** and **Meta data**.
+
+To filter your data:
+
+1. Select a layer from the **Layers window**.
+
+ 
+
+2. Open the **Data filters window** by clicking the tab at the bottom of the screen.
+
+ 
+
+3. Add a filter to the **Data filters window** by right-clicking the window and then selecting **Add filter** from the context menu.
+
+4. Select a field, select an operation and enter a value. For example, to exclude all peaks with an intensity of less than 6999, set **field** to **Intensity**, **operation** to **=>** and set the value to 7000. Click **Ok** on the panel to apply the changes.
+
+ 
+
+5. You should see only see data that satisfies the specified criteria.
+
+### Additional topics
+
+You might want to check out the following topics:
+
+```{toctree}
+:maxdepth: 1
+
+visualize-with-openms/views-in-toppview.md
+visualize-with-openms/hotkeys-table.md
+```
diff --git a/docs/tutorials/TOPP/hotkeys-table.md b/docs/openms-applications-and-tools/visualize-with-openms/hotkeys-table.md
similarity index 100%
rename from docs/tutorials/TOPP/hotkeys-table.md
rename to docs/openms-applications-and-tools/visualize-with-openms/hotkeys-table.md
diff --git a/docs/tutorials/TOPP/views-in-toppview.md b/docs/openms-applications-and-tools/visualize-with-openms/views-in-toppview.md
similarity index 100%
rename from docs/tutorials/TOPP/views-in-toppview.md
rename to docs/openms-applications-and-tools/visualize-with-openms/views-in-toppview.md
diff --git a/docs/tutorials-and-quickstart-guides/tutorials.md b/docs/tutorials-and-quickstart-guides/tutorials.md
index 5db0e369..7b4b2799 100644
--- a/docs/tutorials-and-quickstart-guides/tutorials.md
+++ b/docs/tutorials-and-quickstart-guides/tutorials.md
@@ -137,10 +137,10 @@ KNIME is available for those who want a graphical application to create and use
Go to the Node repository
In the search field, type Input File. You should see an Input File node in the filtered list.
Drag and drop the Input File node from the Node repository into the workspace.
-  +
+  Repeat steps b and c for the FileInfo node and the Output File node.
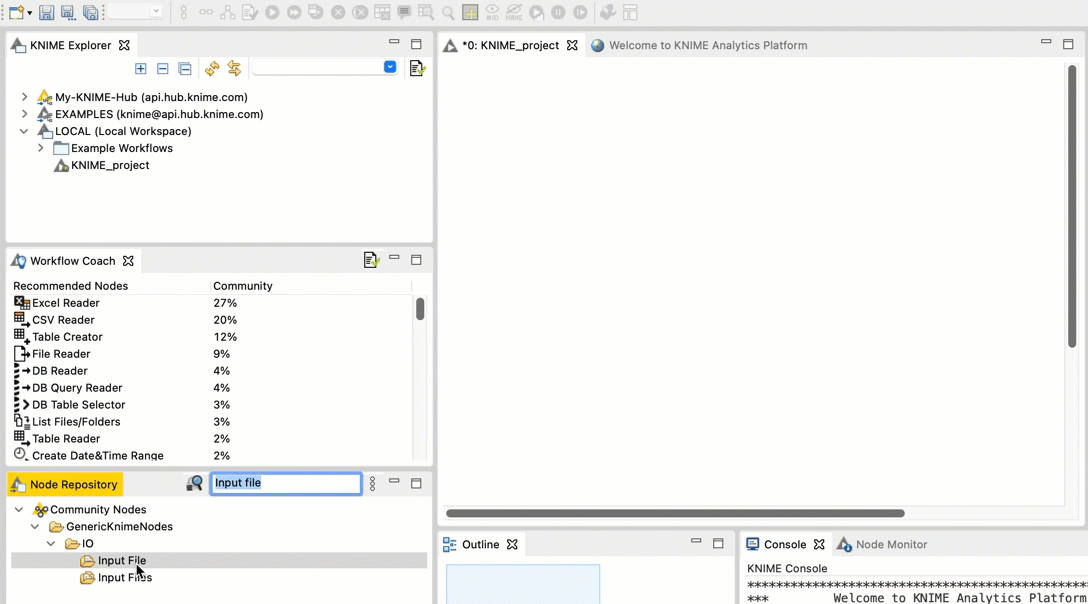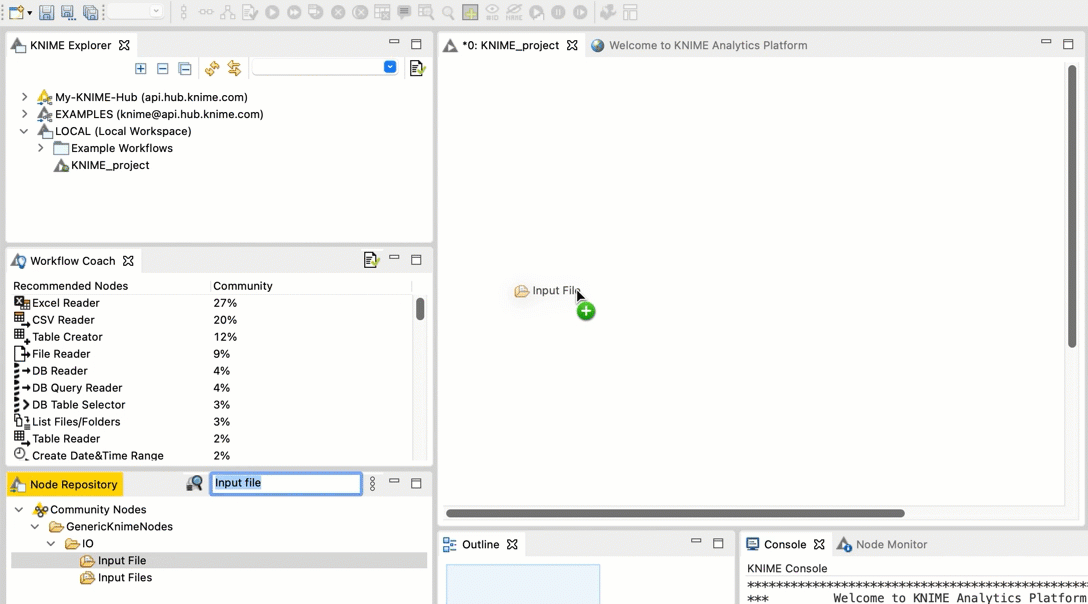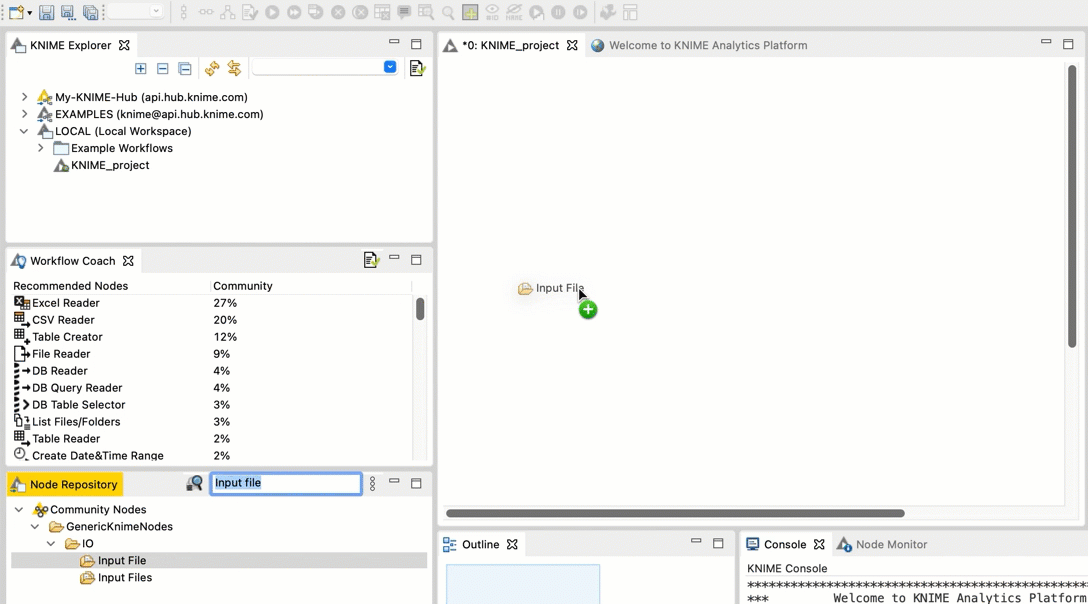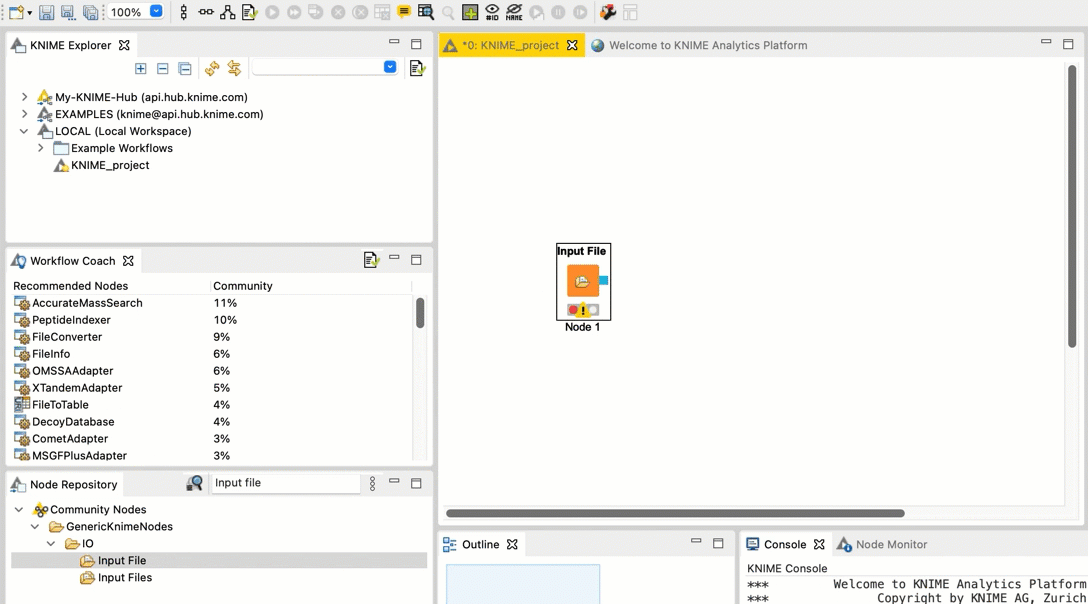
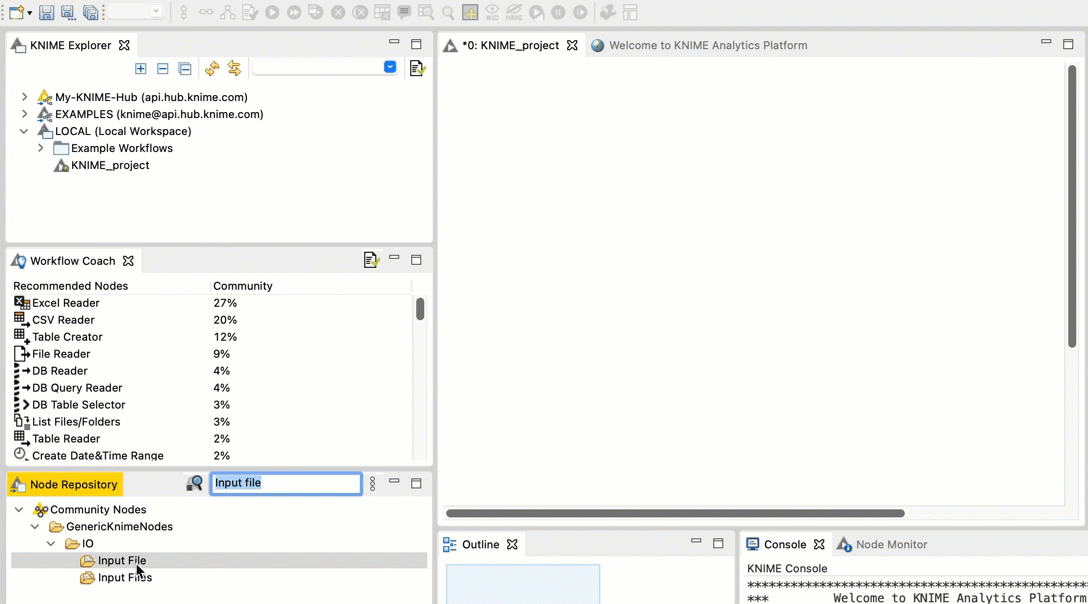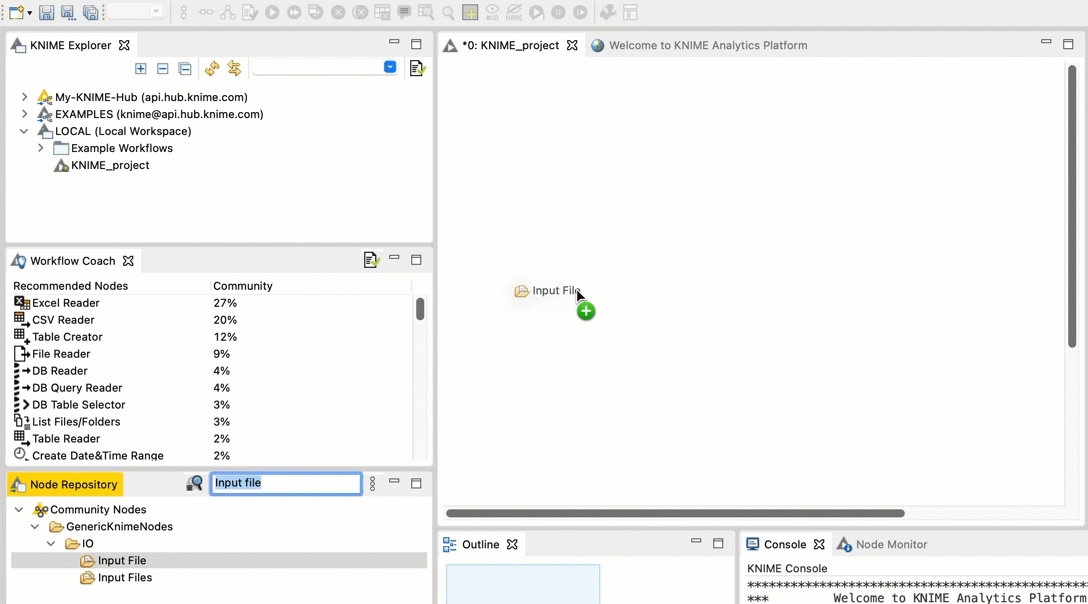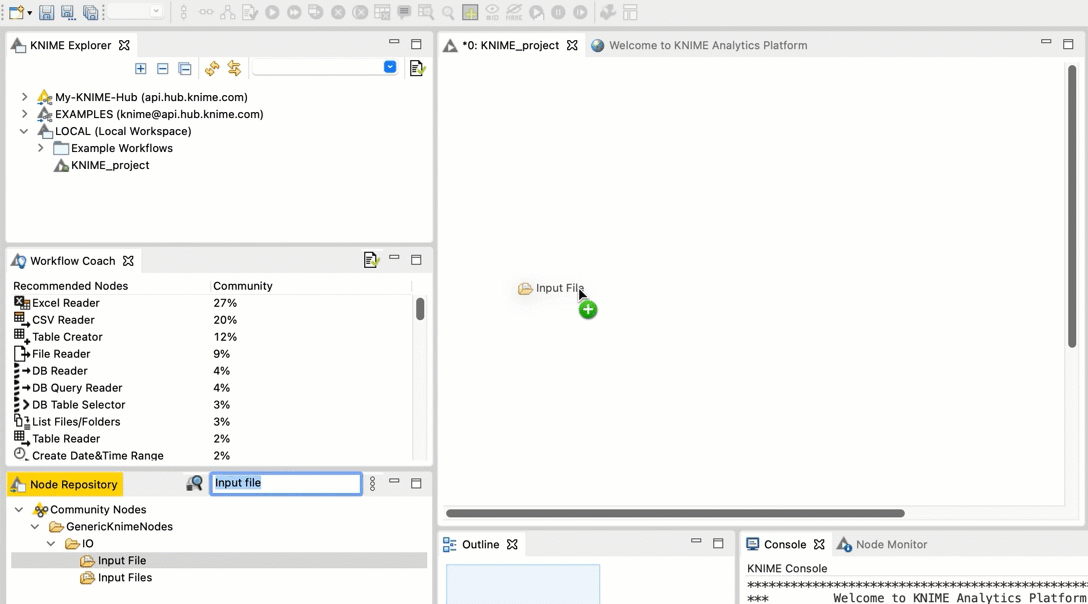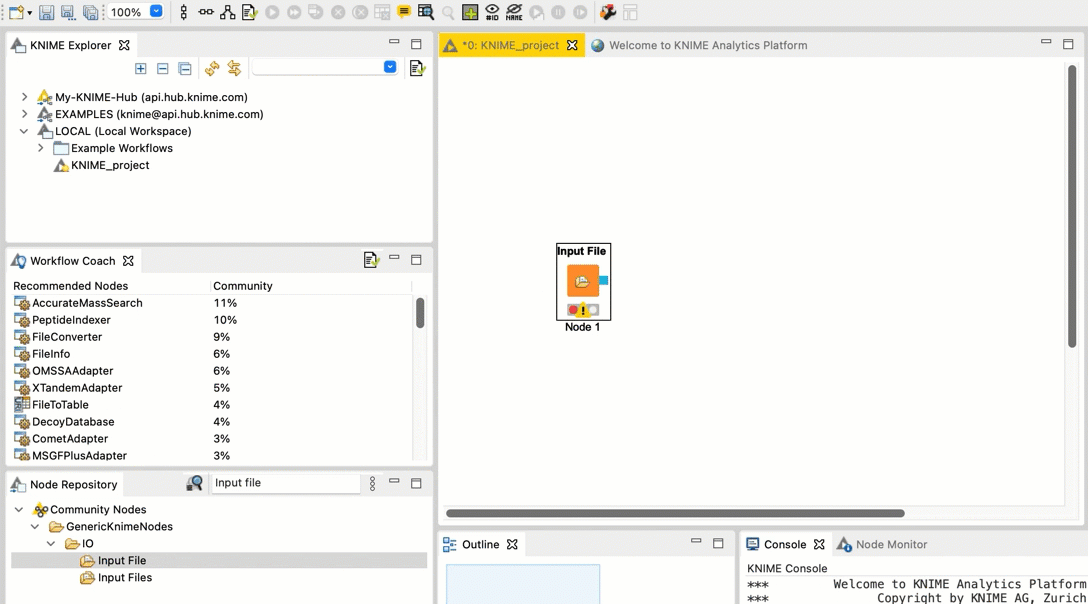
Connect the Input File node to the FileInfo node. Connect the FileInfo node to the Output File node. Your workflow should look like this:
-
Repeat steps b and c for the FileInfo node and the Output File node.
Connect the Input File node to the FileInfo node. Connect the FileInfo node to the Output File node. Your workflow should look like this:
- 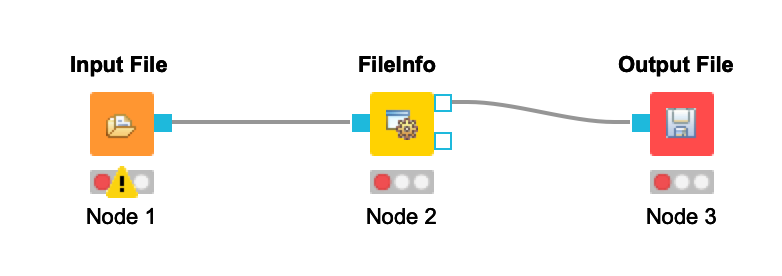 +
+ 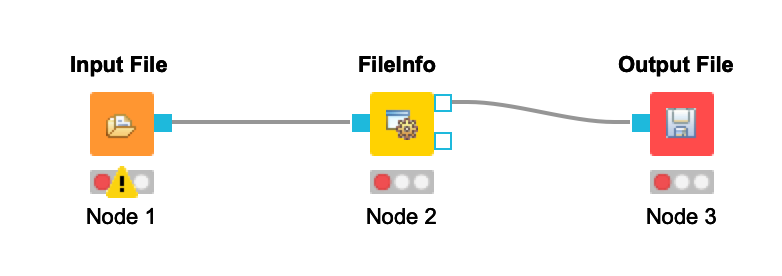 Once you configure your nodes in the next step, the warnings should disappear.
@@ -153,12 +153,12 @@ KNIME is available for those who want a graphical application to create and use
Either choose an existing file or enter the file name of the new file. If you choose an existing file, select the Overwrite file(s) if it/they exist checkbox.
Click OK.
Your workflow should be cleared of warnings and look like this:
-
Once you configure your nodes in the next step, the warnings should disappear.
@@ -153,12 +153,12 @@ KNIME is available for those who want a graphical application to create and use
Either choose an existing file or enter the file name of the new file. If you choose an existing file, select the Overwrite file(s) if it/they exist checkbox.
Click OK.
Your workflow should be cleared of warnings and look like this:
- 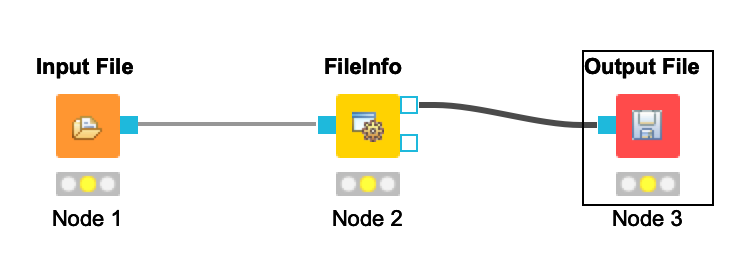 +
+ 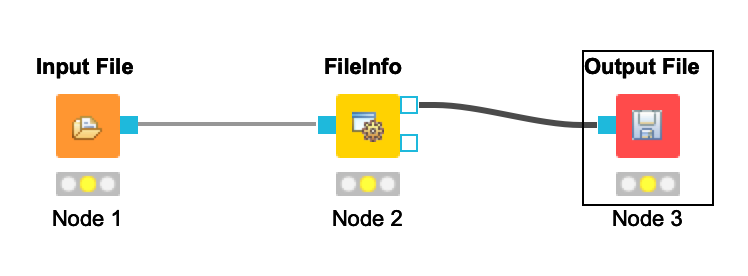 -
+
**5. Play the workflow.**
-
+
**5. Play the workflow.**
- - Click the Execute all executable nodes
 button in the toolbar at the top of the screen.
button in the toolbar at the top of the screen.
+ - Click the Execute all executable nodes
 button in the toolbar at the top of the screen.
button in the toolbar at the top of the screen.
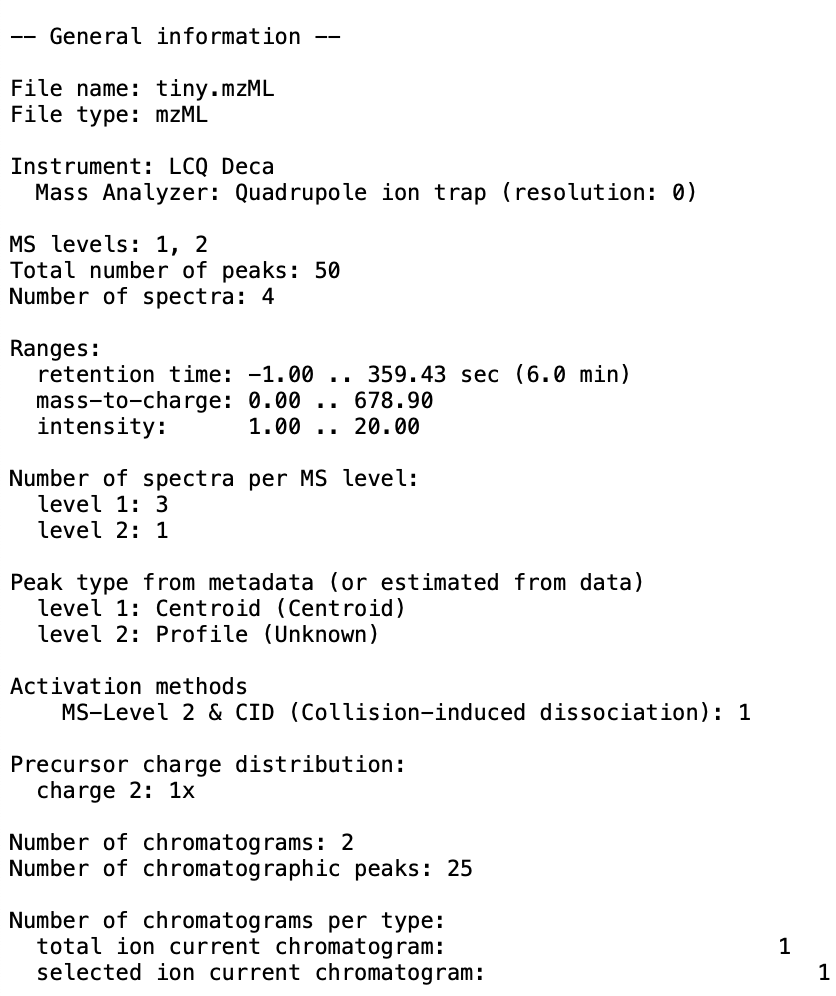
- You should produce an output file at the specified location. Here is an example of what can be produced:
- 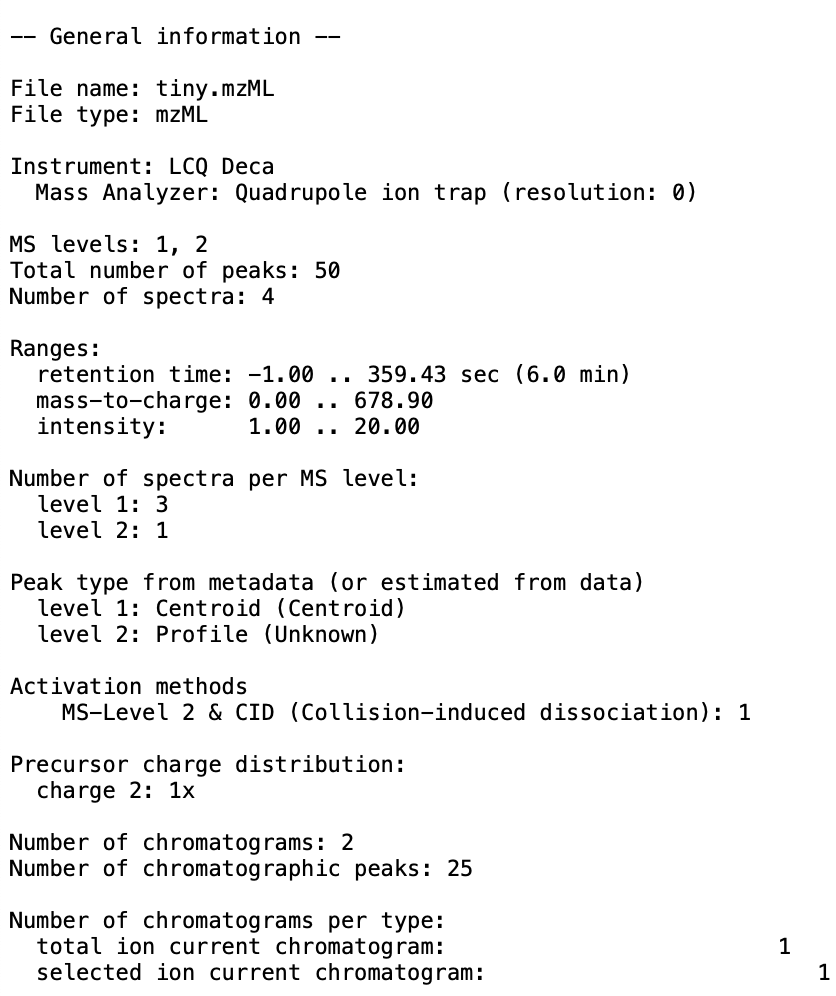 +
+ 
 +
+ 
 +
+  Once you configure your nodes in the next step, the warnings should disappear.
@@ -153,12 +153,12 @@ KNIME is available for those who want a graphical application to create and use
Once you configure your nodes in the next step, the warnings should disappear.
@@ -153,12 +153,12 @@ KNIME is available for those who want a graphical application to create and use
 +
+  -
+
**5. Play the workflow.**
-
+
**5. Play the workflow.**
 button in the toolbar at the top of the screen.
+
button in the toolbar at the top of the screen.
+  button in the toolbar at the top of the screen.
button in the toolbar at the top of the screen. +
+ 