-
Notifications
You must be signed in to change notification settings - Fork 57
Description
Hi there
My name is Antonio and I am studying Bioinformatics at University of Copenhagen.
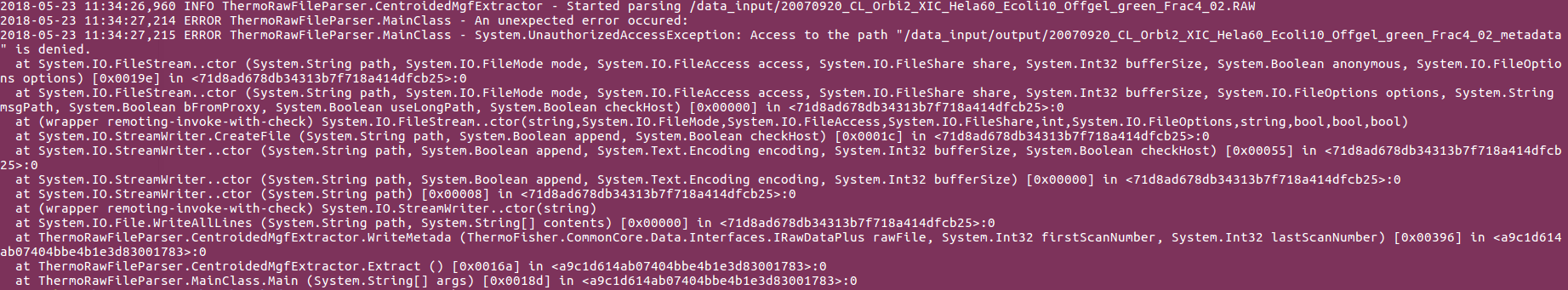
I was directed to this tool by @Maux82 in CompOmics/moFF#26. I am trying to use ThermoRawFileParser through the Docker image and I am encountering some path issues when mapping the Docker volumes to files in my local filesystem.
I built the image using the command in your README.md
docker build --no-cache -t thermorawparser .
Then I tried running
docker run -v $DATA_INPUT:/data_input -i -t --user biodocker thermorawparser mono /home/biodocker/bin/bin/Debug/ThermoRawFileParser.exe -i=/data_input/$RAW_FILE -o=/data_input/output -m -c=$PXD
where
- DATA_INPUT is an absolute path to the directory where my RAW_FILE is
- RAW_FILE is the name of my raw file (no path prepended, i.e myfile.RAW)
- PXD is a custom string.
I get the following error (please click on the image to get the full picture):
The file I am trying to convert is available here:
https://www.ebi.ac.uk/pride/data/archive/2014/09/PXD000279/20070920_CL_Orbi2_XIC_Hela60_Ecoli10_Offgel_green_Frac4_02.RAW
and it belongs to the dataset used in the MaxLFQ paper http://www.mcponline.org/content/13/9/2513.long
I am running this on an Ubuntu 16.04.
Could it be due to the fact that the path /data_input does not exist inside the container filesystem?
Thanks beforehand for your help and for developing such a useful tool!
Best
Antonio