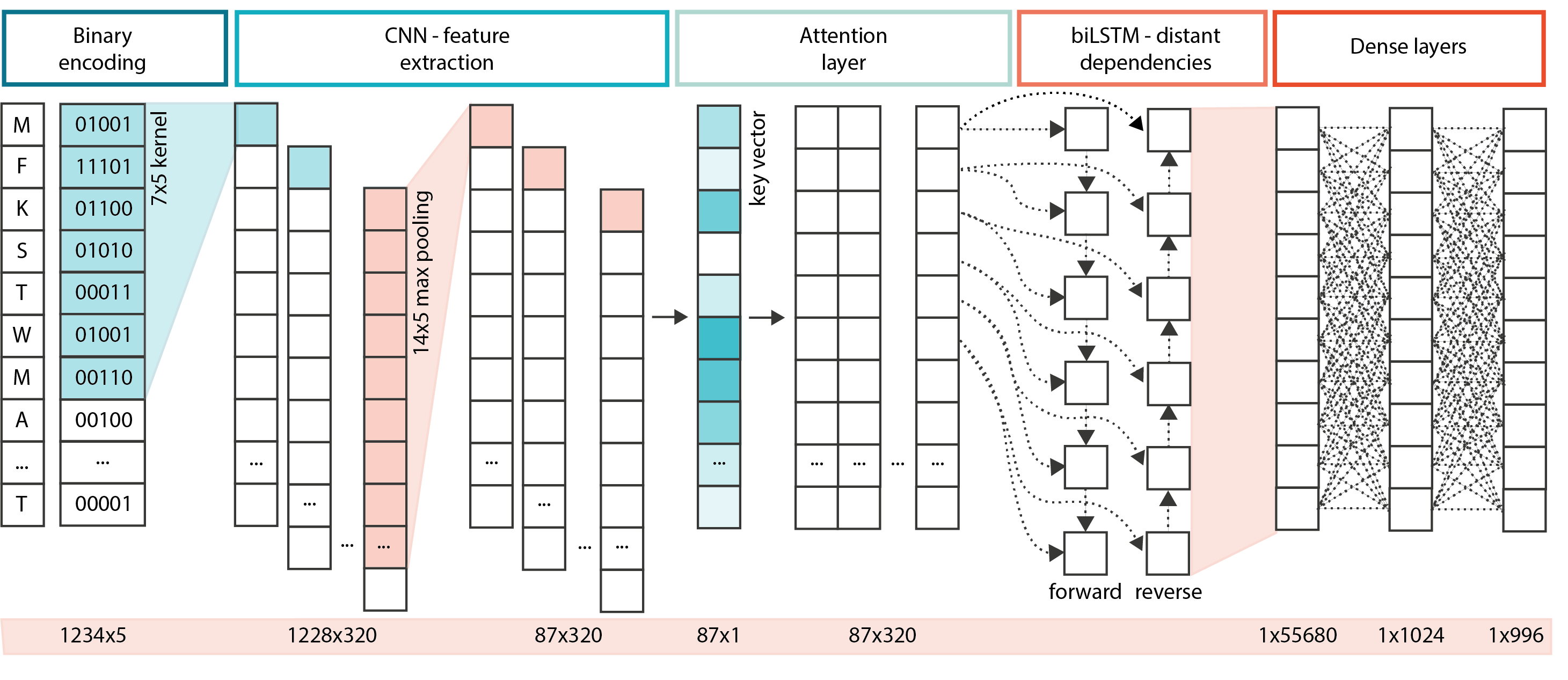
PF-NET is a multi-layer neural network, consisting of a CNN, attention layer, and a biLSTM, that accurately annotates sequences from 996 protein families. An online application of PF-NET can be found at https://sozzanilab.shinyapps.io/PF-NET_Shiny/
Van den Broeck, L., Bhosale, D.K., Song, K. et al. Functional annotation of proteins for signaling network inference in non-model species. Nat Commun 14, 4654 (2023). https://doi.org/10.1038/s41467-023-40365-z
- Python 3.6.12
- Numpy
- Pickle
- Sklearn
- Tensorflow 2.3.1
- Keras 2.4.0
We selected 996 protein families from Pfam (https://pfam.xfam.org/), focusing on protein families within the plant and animal kingdom. We extracted accompanying sequences from Pfam's underlying sequence database using the following tables: pfama_reg_full_significant and pfamseq. A third table (pfamnn) was created that contains the 996 protein families. MySQL scripts to generate the pfamnn table and to extract sequences are located in Src/MySQL.
All extracted sequences were preprocessed by retaining unique sequences and appending labels in case when multiple labels per sequence were present. Sequences with a length < 1234 amino acids were kept (source code: "Src/Preprocessing/select-sequences-labels", extracted sequences: "Data/extracted-pfam-total-data/aa-sequences-crop.npz" and corresponding labels: "Data/extracted-pfam-total-data/pfam-labels-crop.npz"). These sequences are subdivided into batches of +/- 1 milion sequences (source code: "Src/Preprocessing/divide-pfam-data-batches"). Labels and sequences were encoded and also subdivided into batches (source code: "Src/Preprocessing/encode-labels-divide-batches" and "Src/Preprocessing/encode-pad-divide-batches-sequences", encoded labels: "Data/extracted-pfam-total-data/encoded-labels.npz"). A stratified dataset was then created with 6 batches for single label sequences (source code: "Src/Preprocessing/create-straitified-dataset"). Sequences with multiple labels were not used for training.
The total training dataset consisted of 7,385,028 sequences, covering the entire tree of life.
The source code for generating training, validation, and testing datasets (save-train-val-test.py), for the model (model.py and loss.py), for training (train.py), testing (predict.py and test-predict.py), and predicting (validation-predict.py) can be found in "Src/Model/".