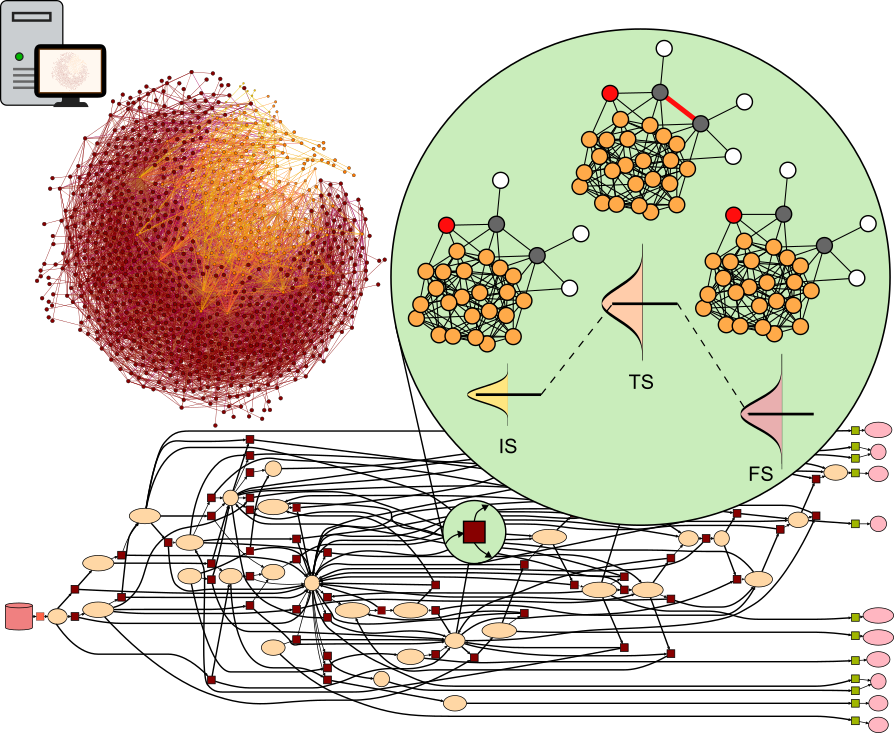
CARE (Catalysis Automated Reaction Evaluator) is a tool for generating and manipulating chemical reaction networks (CRNs) on catalytic surfaces. CARE is powered by data-driven models such as GAME-Net-UQ, Open Catalyst models, MACE, etc.
We recommend installing care-crn from PyPI. A developer installation is also available for those who wish to contribute.
This is the fastest way to get care-crn and its core dependencies.
pip install care-crncare-crn interfaces with several external ML Interatomic Potentials (MLIPs). These must be installed separately.
You can install the Python wrappers for these evaluators using pip's "extras" syntax.
-
For OCP (FAIRChem-v1): First, install
torch_sparseandtorch_scatterby following the instructions on the PyTorch Geometric page. Then, run:pip install care-crn[ocp]
-
For other evaluators: You can install MACE, PET-MAD, Orb, and SevenNet by running:
pip install care-crn[mace] pip install care-crn[petmad] pip install care-crn[orb] pip install care-crn[sevennet]
Or all at once:
pip install care-crn[ocp,mace,petmad,orb,sevennet]
NOTE: There currently is a dependency clash during installation of OCP and MACE evaluators related to the
e3nnlibrary (see: this issue for MACE). Installation might result in an incompatibility warning, but both evaluators should work correctly if the installation order shown above is followed.
To run microkinetic simulations with Julia, install it and the required packages:
# Install Julia and add version 1.11
curl -fsSL https://install.julialang.org | sh -s -- --yes && ~/.juliaup/bin/juliaup add 1.11
# Add DifferentialEquations.jl and LinearSolve.jl
julia -e 'import Pkg; Pkg.add("DifferentialEquations"); Pkg.add("LinearSolve");'⏲ Julia setup time estimate: ~13min (Ubuntu), ~9min (macOS)
If you want to contribute to the code or use the very latest (unstable) version, you can install from the source.
- ⏲ Total installation time estimates: ~18min (Ubuntu), ~11min (macOS).
- 💾 Required disk space: ~6.5 GB (Conda environment), ~4.3 GB (Julia+dependencies)
-
Clone the repo:
git clone git@github.com:LopezGroup-ICIQ/care.git cd care -
Create a conda environment:
conda create -n care_env python==3.12 conda activate care_env
-
Install the package in "editable" mode: (The
-eflag links the installation to your source code)python3 -m pip install -e .NOTE: macOS users might need to launch a new shell at this point in order for the entry points to work correctly.
-
Install optional dependencies: (Note the syntax is slightly different from the PyPI install)
python3 -m pip install -e .[ocp] python3 -m pip install -e .[mace] # etc.
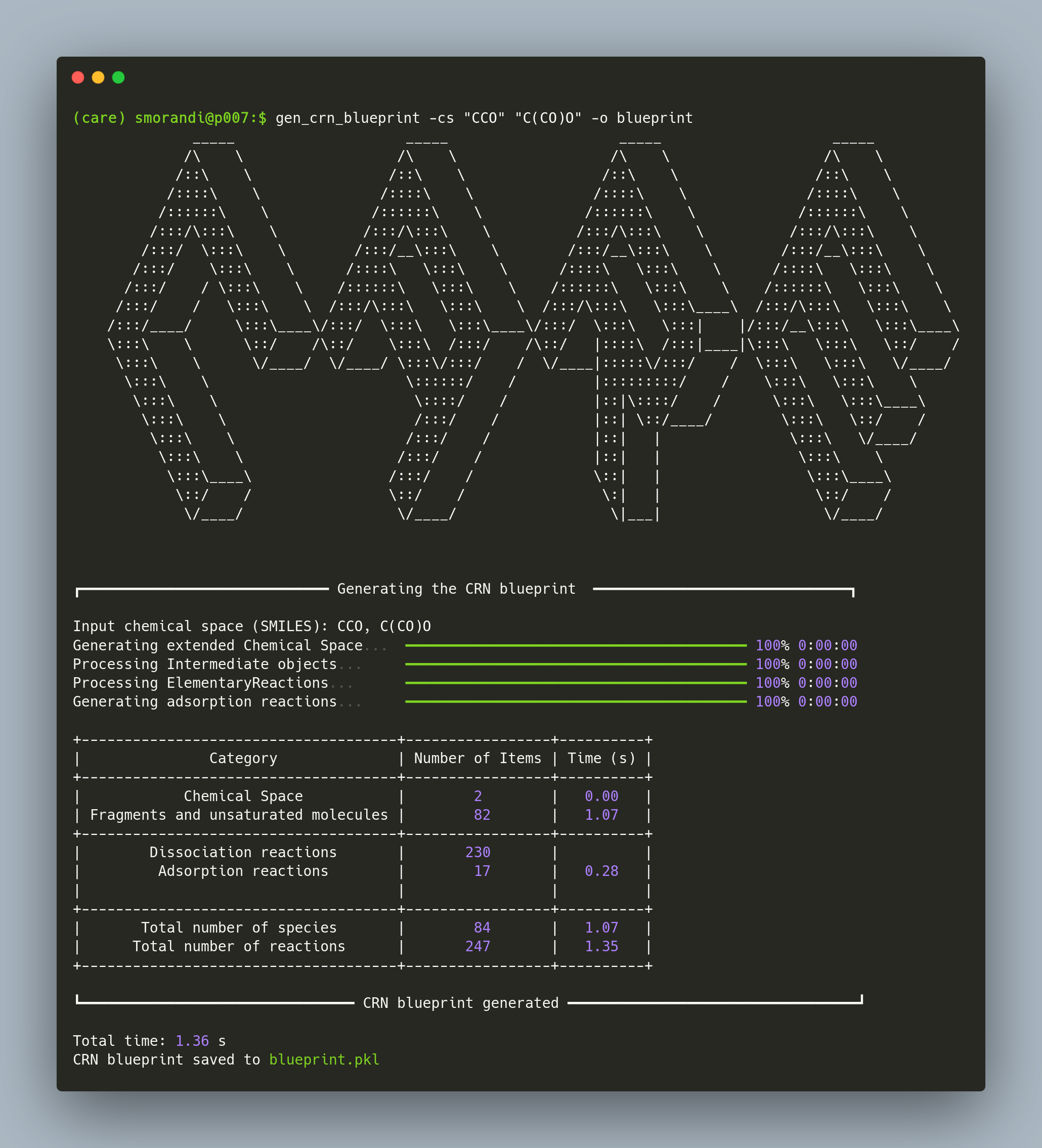
The blueprint can be constructed in two ways, by providing (i) the network carbon and oxygen cutoffs ncc and noc, or (ii) the chemical space as list of SMILES.
gen_crn_blueprint -h # documentation
gen_crn_blueprint -ncc 2 -noc 1 -o output_name # Example from ncc and noc
gen_crn_blueprint -cs "CCO" "C(CO)O" -o output_name # Example from user-defined chemical spaceThe CRN blueprint is stored as pickle file. To access the blueprint, do:
from pickle import load
with open('path_to_blueprint_file', 'rb') as f:
intermediates, reactions = load(f)The range of catalyst materials on which CRNs can be constructed depends on the domain of the data-driven energy evaluator employed to derive the reaction properties. Currently, CARE provides interfaces to GAME-Net-UQ, OCP models, and MACE-MP potentials.
eval_crn -h # documentation
eval_crn [-i INPUT] [-bp BP] [-o OUTPUT] [-ncpu NUM_CPU]This script requires an input toml file defining the material/surface of interest, the model of choice and its settings. The output is a ReactionNetwork object stored as pickle file. You can find examples of input files here.
For macOS we noticed a lower performance in the CRN generation due to Python multiprocessing (see Contexts and start methods in the documentation)
run_kinetic [-i INPUT] [-crn CRN] [-o OUTPUT]This script runs microkinetic simulation starting from the evaluated reaction network and an input toml file defining the reaction conditions, solver, inlet conditions. The results are stored as a pickle object file.
You can run the entire pipeline (blueprint generation ➡ energy evaluation ➡ kinetic simulation) running the care_run script:
care_run -h # documentation
care_run -i input.toml -o output_nameThis will generate a directory output_name containing a crn.pkl with the generated reaction network.
Examples of input .toml files can be found here.
We currently provide three tutorials, available in the notebooks directory:
- CRN generation and manipulation
- Energy evaluator interface implementation
- Microkinetic simulations
- Adsorbate placement
The code is released under the MIT license.
- A Foundational Model for Reaction Networks on Metal Surfaces Authors: S. Morandi, O. Loveday, T. Renningholtz, S. Pablo-García, R. A. Vargas Hernáńdez, R. R. Seemakurthi, P. Sanz Berman, R. García-Muelas, A. Aspuru-Guzik, and N. López DOI: 10.26434/chemrxiv-2024-bfv3d