-
Notifications
You must be signed in to change notification settings - Fork 1.4k
Split patch functionality for PatchWSIReader
#4251
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
Signed-off-by: Behrooz <3968947+drbeh@users.noreply.github.com>
Signed-off-by: Behrooz <3968947+drbeh@users.noreply.github.com>
|
I lose the overall picture of the WSI-related reader/dataset modules, would be great to have some tutorials in https://github.com/Project-MONAI/tutorials/tree/master/modules to demonstrate the use cases, otherwise it's not easy to help review these PRs. |
Typically we would want to decouple behaviours so that we have the option to choose what to use, and reuse functionality elsewhere. Using |
|
(For an example of the tutorials: this PR proposes code changes #4236, and the corresponding usage is shown in a PR to the tutorials Project-MONAI/tutorials#690. These greatly help the reviewing/discussion process...) |
|
@Nic-Ma @ericspod, this split does not have any randomness (like what we have discussed before) and I had in mind to use the transform only but the reason to have the split here is to managing the following transforms. What I mean is that the split transform generate several patches (from few patches to few hundreds or thousands of patches), and we want these patches treated independently like images in a batch. It was not clear to me how its output will be handle by its following transforms. Assume the following:
I would really like to make this as a separate transform so I'd appreciate it if you have any solution that can create the batches as we like (without implementing a new collate function). |
|
Hi @drbeh , I think your problem is similar to our Thanks. |
@Nic-Ma, this is what the current implementation is already leveraging and that's why I put However, this is not the case if a transform at the middle of chain creates a list of images, right? |
I think the problem is similar, so how this is being batched? Is there any issue for the other transforms if it is a list? |
|
Thanks. |
Signed-off-by: Behrooz <3968947+drbeh@users.noreply.github.com>
Signed-off-by: Behrooz <3968947+drbeh@users.noreply.github.com>
Signed-off-by: Behrooz <3968947+drbeh@users.noreply.github.com>
|
@Nic-Ma @ericspod, I have updated the @wyli, I have updated the tumor detection pipeline that uses these transforms and dataset: Project-MONAI/tutorials#697 |
Signed-off-by: Behrooz <3968947+drbeh@users.noreply.github.com>
 Nic-Ma
left a comment
Nic-Ma
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
thanks, @Nic-Ma could you please help run and verify the tutorial (perhaps with a subset of CAMELYON16 for a quick setup)? |
That's a good idea. I'll add a subset dataset to the PR of the tutorial to facilitate the testing but we can remove it before merge. |
Signed-off-by: Behrooz <3968947+drbeh@users.noreply.github.com>
|
@Nic-Ma @wyli, I have added a subset dataset ('training.txt') into this tutorial PR: Project-MONAI/tutorials#697
Please let me know if you have any question. |
|
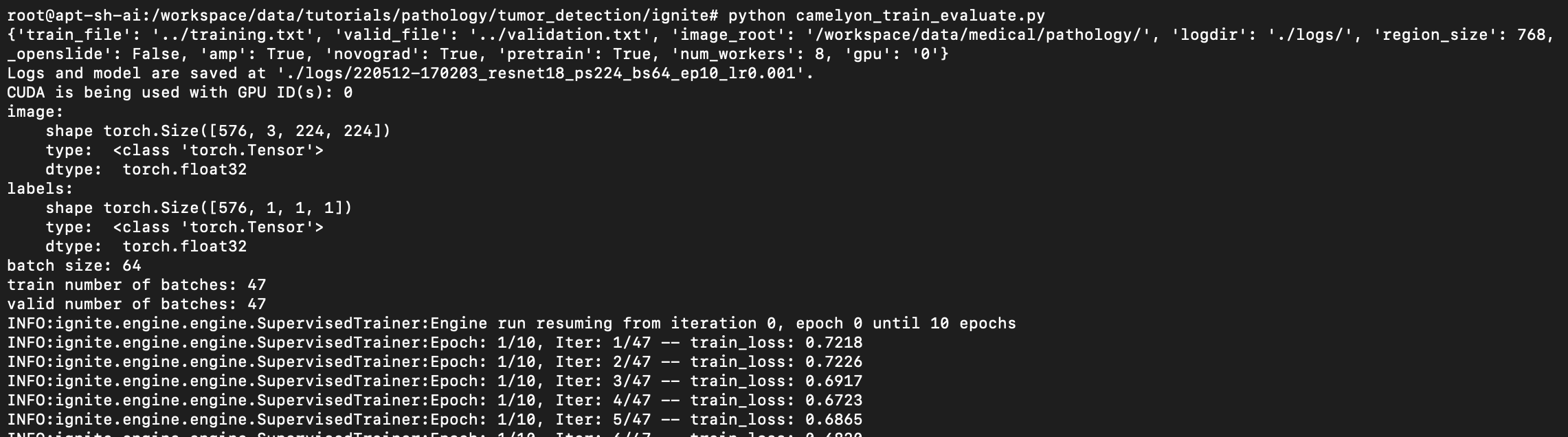
Hi @drbeh @wyli , I tried to run the tutorial, seems it can run normally: Thanks. |
Signed-off-by: Behrooz <3968947+drbeh@users.noreply.github.com>
|
/build |
Fixes #4247
Description
This PR add split patch functionality to
PatchWSIReader.Status
Ready
Types of changes
./runtests.sh -f -u --net --coverage../runtests.sh --quick --unittests --disttests.