-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
[MRG+1] Add function for random parcellation #4955
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
Codecov Report
@@ Coverage Diff @@
## master #4955 +/- ##
==========================================
+ Coverage 87.85% 87.87% +0.01%
==========================================
Files 353 353
Lines 62988 63177 +189
Branches 10783 10811 +28
==========================================
+ Hits 55337 55515 +178
- Misses 4909 4913 +4
- Partials 2742 2749 +7 |
mne/label.py
Outdated
| Name of the subject as in SUBJECTS_DIR. | ||
| n_parcel : int | ||
| Total number of cortical parcels | ||
| hemis : array | int |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
it should be called hemi and be a string. Following the API of pysurfer:
https://github.com/nipy/PySurfer/blob/master/surfer/viz.py#L301
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I used the doc of the function "grow_labels". If need to change it, probably should do it for grow_labels too.
mne/label.py
Outdated
| """Random cortex parcellation.""" | ||
| labels = [] | ||
| parcel_size = np.floor((len(vertices_[hemis[0]]) + | ||
| len(vertices_[hemis[1]])) / n_parcel) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think
len(vertices_[hemis[0]]) + len(vertices_[hemis[1]])) // n_parcel
is nicer and is the same
mne/label.py
Outdated
|
|
||
| # prepare parcellation | ||
| parc = np.empty(n_vertices, dtype='int32') | ||
| parc[:] = -1 |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
mne/label.py
Outdated
| parc[:] = -1 | ||
|
|
||
| # initialize active sources | ||
| s = 0 |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
line is useless
mne/label.py
Outdated
| name = 'label_' + str(i) | ||
| label_ = Label(vertices, hemi=hemi, name=name, subject=subject) | ||
| labels.append(label_) | ||
| parc_both += [parc] |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
is this the same algo as in brainstorm?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I followed the same logic as brainsotm code:
- define number of vertices per parcel
- grow parcels till there are no free neighbor vertices or till riches max size
- merge smallest parcels till rich the needed number
mne/label.py
Outdated
|
|
||
|
|
||
| def random_parcellation(subject, n_parcel, hemis, subjects_dir=None, | ||
| surface='white'): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
you should use np.random and have a random_state parameter to make the result reproducible.
git grep random_state
to see how we do it.
|
for the lazy curious is the output of the script shared by @makkostya |
|
Oh the shame of having to run the example yourself to produce this output. It should be done by CircleCI and sphinx-gallery :) |
|
I used the doc of the function "grow_labels". If need to change it,
probably should do it for grow_labels too.
hum indeed. I would recommend to use the surfer way and we'll deprecate the
hemis param in grow_label in another PR.
Having to pass array of int is really not pythonic
|
|
Ok, I'll change it. And can do PR for "grow_labels" too |
|
thanks a lot
|
grow_label function has parameter hemis which is different from pysurf because we can associate the hemisphere to each source seed. So may be it makes sense to keep it like it is? |
mne/label.py
Outdated
| parc = np.full(n_vertices, -1, dtype='int32') | ||
|
|
||
| # initialize active sources | ||
| rng = check_random_state(random_state) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I would have put the check_random_state outside of the for loop on hemi no?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, done.
|
LGTM any other review? |
|
LGTM but @makkostya your |
|
@larsoner , I added my |
|
Yep all better, thanks @makkostya |
* Add functions to create random parcellation * add test for random_parcellation * add random_parcellation to python_reference * fixe pep8 * fixe pep8 mne-tools#2 * fixe pep mne-tools#3 * add random_state, small corrections * fixes random_state * FIX: Alphabetical
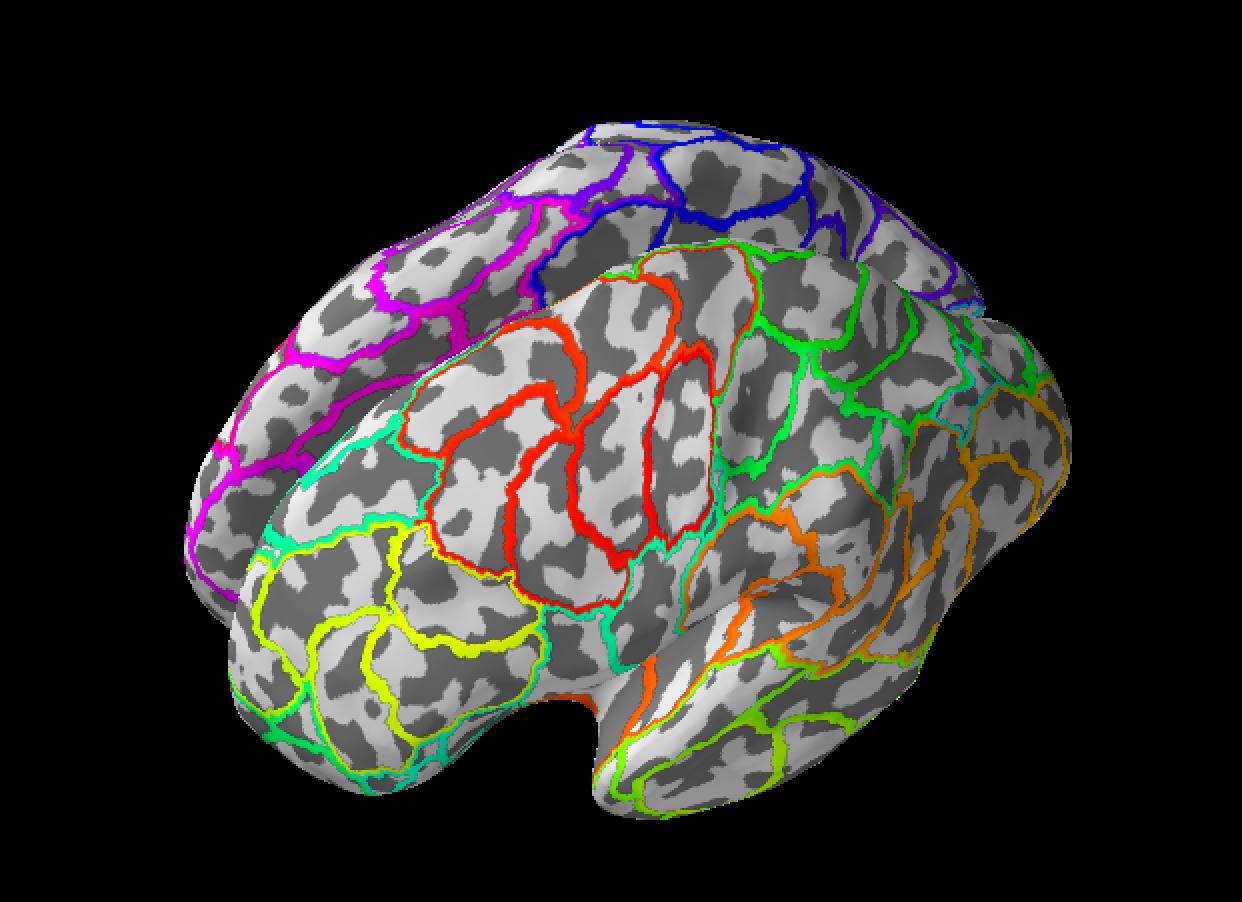
@agramfort @massich @mathurinm
This PR add the new function to produce random parcellation of the cortex. It grows a number (input parameter) of random not intersecting cortical regions which cover the whole surface.
Here is example of use (you should change annotation file names):