-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
[WIP] Add CSD whitening to DICS beamformer #7021
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
 larsoner
left a comment
larsoner
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Looks like a great start, love to see a small diff bring a nice enhancement. (I know it's not done yet but it looks close!)
|
|
||
| # Set picks, use a single sensor type | ||
| picks = mne.pick_types(raw.info, meg='grad', exclude='bads') | ||
|
|
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hooray!
mne/time_frequency/csd.py
Outdated
| from ..cov import Covariance # to avoid circular import | ||
| return Covariance(data, self.ch_names, bads=[], projs=self.projs, | ||
| nfree=self.n_fft) | ||
| return None |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
debugging cruft?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
probably :)
 britta-wstnr
left a comment
britta-wstnr
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hooray for combining sensors for DICS!
I fear I came to question leadfield whitening and its effect on DICS with multiple frequencies - see my reviewing comments ... What is your opinions on this @larsoner @wmvanvliet ?
| computing the inverse. | ||
| noise_csd : instance of CrossSpectralDensity | None | ||
| Noise cross-spectral density (CSD) matrices. If provided, whitening | ||
| will be done. The noise CSDs need to have been computed for the same |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The noise CSD matrices need to be computed
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
My reasoning with the grammar was that as you are supplying the CSDs are a parameter to this function, they already have been computed.
mne/beamformer/_dics.py
Outdated
|
|
||
| _, _, proj, vertices, G, _, nn, orient_std = _prepare_beamformer_input( | ||
| info, forward, label, pick_ori, noise_cov=noise_csd_freq, | ||
| combine_xyz=combine_xyz, exp=exp) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Does this really need to be done per frequency now? It was outside the loop before if I see it right.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Oh wait ... we now whiten each lead field differently, is that it? Hummmmm, is that something we want (I do understand why we have it)? Different lead fields per frequency ... ? It strikes me as an odd thing to have. One step further: do we actually have to whiten the lead field or would it be sufficient to whiten the data covariance /CSD matrix.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
We absolutely have to whiten per frequency. We have a different CSD at each frequency, and thus a different beamformer at each frequency. It's just a convenience feature that we pack multiple CSDs into one object and multiple beamformers in a single datastructure.
However, there is now a lot of redundant work happening within _prepare_beamformer_input. I plan to refactor that function to unpack the different things it does.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Well I do understand we get different weights at each frequency. The question is whether we want to have different lead fields, too. Probably not. And that raises the general question why we whiten lead fields - we do not invert the lead field in beamforming as compared to MNE solutions.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I don't know what happens if the lead field is not whitened - if it mathematically would still pan out. Maybe worth a try! AFAIK there is no literature on lead field whitening with beamformers, only covariance matrices - but I might be missing that paper ;)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
And that raises the general question why we whiten lead fields - we do not invert the lead field in beamforming as compared to MNE solutions.
At least for MNE any linear operator you apply to the data you should apply to the lead fields because otherwise the results will be biased. I assume the same would hold for beamformers, no?
For example if you apply a projector to the data, you apply it to the lead fields. If you know that this is the correct thing mathematically to do for beamformers when you apply a projection operation to the data, then it is probably mathematically also the correct thing to do in the case of whitening as well. (The whitener implicitly also applies the projection matrix FWIW.)
To me it's not so much about having "different lead fields" as it is about applying the same operators to data and the lead fields before doing computations on them. But maybe I'm thinking about it the wrong way...
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Well, it is def. the case that whatever is done on the original data needs to be done on the lead field as well. However - does the data need to be whitened to take care of scaling or is it enough to do the covariance matrix?
I quickly discussed things with @sarangnemo and he shares my intuition about this maybe not being ideal. I will try to run some data through differently whitened beamformers and report back!
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
in the DICS code, there is no data, there is only the CSD matrix.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Co-Authored-By: Eric Larson <larson.eric.d@gmail.com>
Codecov Report
@@ Coverage Diff @@
## master #7021 +/- ##
=========================================
Coverage ? 89.76%
=========================================
Files ? 444
Lines ? 79430
Branches ? 12699
=========================================
Hits ? 71297
Misses ? 5349
Partials ? 2784 |
Co-Authored-By: Eric Larson <larson.eric.d@gmail.com>
|
Azure style is real: Looks like all beamformer tests fail: So far the example looks perhaps a tiny bit better than https://mne.tools/dev/auto_examples/inverse/plot_dics_source_power.html Great to see this progress @wmvanvliet ! |
|
So there is no big difference in this case whether we whiten the lead field or not? I guess then the sparse thing to do, also in accordance with beamformer literature (Sekihara et al., 2008 https://ieeexplore.ieee.org/document/4454055 ), is to drop whitening of the lead field and data. cc @sarangnemo |
|
I have tested the whitening also on some other data we have internally (cannot share) that is more noisy and with smaller effect sizes. For that case, whitening the leadfield "explodes" the result. Not whitening the leadfield gives sensible results. Evidence is accumulating against whitening the leadfield... |
|
... I still don't completely understand why whitening the leadfield would be bad in beamforming but good in MNE. Could it be because MNE consideres the leadfield "as a whole", while a beamformer only operates on 3 columns at a time? |
|
is the whitener consistently applied to every object? data cov, CSD
matrices, noise cov and leadfield?
if you do a simulation with some random coloring the pipeline
with whitening should be invariant.
I would do something like this to check the implementation as I don't
understand why it may be
a bad idea to whiten
… |
|
What do you mean by "simulation with some random coloring"? I agree that this implementation needs to be tested. I think I apply whitening constently to all objects, but we need to make sure. |
|
apply a full rank random matrix to the data and the leadfield and see if it
affects the results. It should not if whitening is properly implemented.
… |
|
ah, I see. Good idea! |
|
I think I finally know where things went wrong! I forgot to whiten the CSD in the Check out this gist that multiplied the first 50 channels by 100 to make them artificially super loud. The whitening neatly undoes this: https://gist.github.com/wmvanvliet/bf27cbf56a176651205789d1507870b6 |
|
you want to MRG this one then? good for review?
|
|
So, @wmvanvliet, @ckiefer0, and me have been discussing this a few times over the last weeks off Github ... Our conclusion from today: we should take the same whitener (i.e., same noise cov matrix) over the different frequencies to prevent having different forward models per frequency (the motivation that kicked this whole discussion off). |
|
sounds reasonable.
|
|
Agreed this seems reasonable. So instead of having |
|
Definitely not ready for merging yet. Lots of work still to be done to whip this into shape API wise. Of course, comments are always welcome! Regarding the noise CSD, it is probably best if it were computed using exactly the same method as the data CSD. API-wise, I was thinking of putting in a check that the same frequencies are defined for both CSDs. Internally, we will average across the frequencies to obtain a single noise CSD. Still some things to think about:
|
This is now addressed in #7656 with a test that you can manually left- and right- |
|
Closing in favor of #7656 |
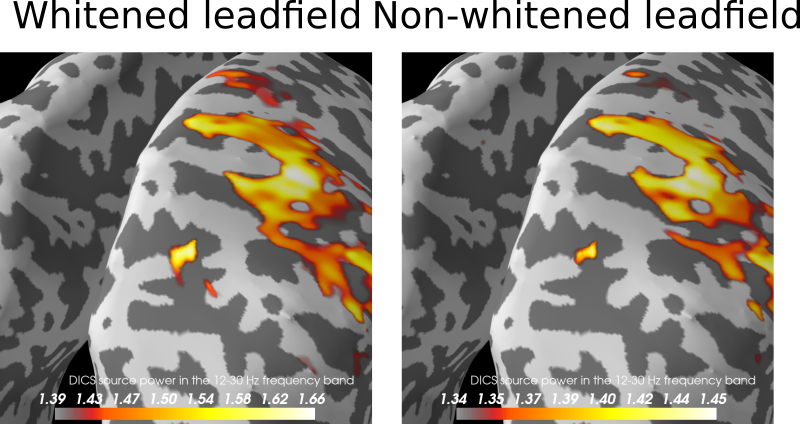
Working on adding whitening support to the DICS beamformer, so we can mix different channel types.
Todo: