-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
artifact detection #7096
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
artifact detection #7096
Conversation
Codecov Report
@@ Coverage Diff @@
## master #7096 +/- ##
=========================================
Coverage ? 89.79%
=========================================
Files ? 447
Lines ? 80506
Branches ? 12873
=========================================
Hits ? 72289
Misses ? 5390
Partials ? 2827 |
|
@AdoNunes can you your file plot_ and not demo_ so it gets executed thanks |
|
@AdoNunes it needs to be in examples/preprocessing folder so it’s rendered by circle ci |
…python into artifact_detection merge origin
sorry, I got it now.... |
|
Why I am getting the ci errors? |
|
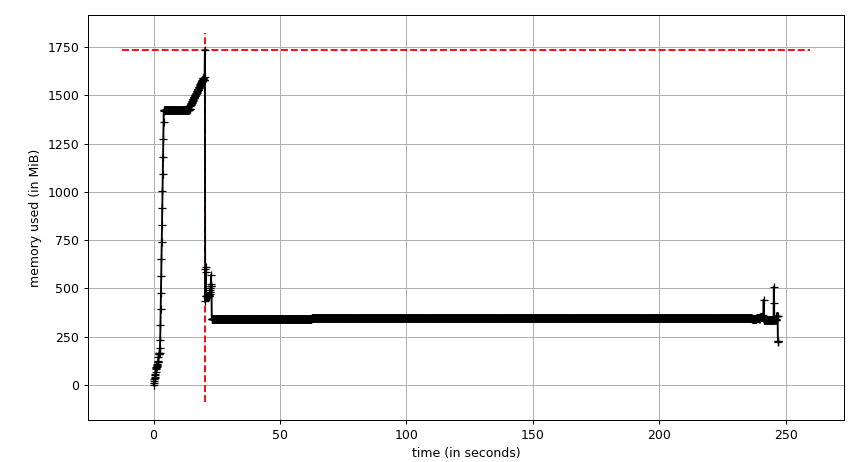
I suspect it's a memory issue. Can you check using a memory profiler how much memory you consume? |
|
To follow what @jasmainak said, you should try to keep examples below 1.5 GB memory consumption, otherwise you risk CircleCI failing. |
…python into artifact_detection conflict Conflicts: examples/preprocessing/plot_artifact_detection.py
|
Try to see if you're making any redundant copies and get rid of them. You can also use |
By shortening even more, changing the order and decimating first then notching it worked :) I will fix the ::doc things and I will resubmit |
|
I am getting an error and I don't know who is wrong. When I crop de raw, say from (100, 300). The raw.times starts at 0, although, the raw.first_time is 100. Is that how it is supposed to be? Because, then the function _calculate_head_pos_ctf returns "quats", the quats[:, 0] is the time, and in chpi.py line 264 -> quats[:, 0] += raw._first_time. So the quats time has an offset that does not match the cropped raw.times and this gives me wrong annotation timings. Any insights? |
|
I don't know about quats but it looks like you just need to handle it correctly by adding raw.first_time in the right places. You might find this tutorial useful: https://mne.tools/stable/auto_tutorials/raw/plot_30_annotate_raw.html Let's make it work so that CircleCI renders something with plots and then we can worry about these details later. |
| raw.notch_filter([60, 120, 180, 240]).resample(300, npad="auto") | ||
|
|
||
| # Detect bad channels | ||
| bad_chns = detect_bad_channels(raw, zscore_v=4, method='both', t1=0, t2=190, |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
bad_chns -> bad_chs to be consistent with rest of repo
mne/artifact_detection.py
Outdated
| decreased local correlation with other channels""" | ||
|
|
||
| # set recording length | ||
| Fs = raw.info['sfreq'] |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Fs -> sfreq
mne/artifact_detection.py
Outdated
| # Get data | ||
| raw_copy = raw.copy().crop(t1x, t2x).load_data() | ||
| raw_copy = raw_copy.pick_types(meg=True, ref_meg=False)\ | ||
| .filter(1, 45).resample(150, npad='auto') |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
You want to hardcode these numbers?
mne/artifact_detection.py
Outdated
| data_chans = raw_copy.get_data() | ||
|
|
||
| # Get channel distances matrix | ||
| chns_locs = np.asarray([x['loc'][:3] for x in |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
chns_locs -> ch_locs
mne/artifact_detection.py
Outdated
| def detect_bad_channels(raw, zscore_v=4, method='both', t1=30, t2=220, | ||
| neigh_max_distance=.035): | ||
| """ detect bad channels based on z-score amplitude deviation or/ and | ||
| decreased local correlation with other channels""" |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Add a docstring to explain what each of these parameters are
mne/artifact_detection.py
Outdated
| """ | ||
| Created on Tue Nov 19 09:40:37 2019 | ||
|
|
||
| @author: an512 |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is not kosher. Put this file under mne/preprocessing/ and then format the header like the other files.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I thought that these scripts were temporary. Fixing it!
mne/artifact_detection.py
Outdated
| """ | ||
|
|
||
| from scipy.stats import zscore | ||
| import mne |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
mne import should come after numpy and scipy
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Actually you shouldn't import mne at all unless you're writing a tutorial or a test. You should do relative imports.
|
@mmagnuski let me know if you're happy now and I'll have a look |
|
oh, I had an old unrefreshed tab it seems :) I don't have anything to add here, but don't have experience with MEG, so I mostly commented on code style and docs. |
|
Do we need a test here is the example enough? |
|
Yes we need some tests. @AdoNunes you can make use of this existing Basically choose some thresholds that should mark segments as bad and verify that they are indeed marked as bad. You can iteratively test pos/velocity translational/rotational since it should have all of these types in a short window (we asked the subject to move their head for this recording) |
| [-0.01964518, 0.13086313, 0.99120578, 0.07258289], | ||
| [0., 0., 0., 1.]]) | ||
|
|
||
| assert_allclose(dev_head_t_ori, dev_head_t['trans'], rtol=1e-5, atol=0) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
All of this needs to be nested inside a function like def test_artifact_detection(): otherwise it will be run on test collection (see how we do it in other files).
|
|
||
| with warnings.catch_warnings(record=True): | ||
| warnings.simplefilter("ignore") | ||
| raw = read_raw_fif(raw_fname, allow_maxshield=True).load_data() |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
allow_maxshield='yes' then you don't need any warnings gymnastics (see existing cHPI code for example)
|
Sure!
…On Tue., Jan. 28, 2020, 1:31 p.m. Eric Larson ***@***.***> wrote:
***@***.**** commented on this pull request.
------------------------------
In examples/preprocessing/plot_artifact_detection.py
<#7096 (comment)>:
> +plt.axhline(y=mean_distance_limit, color='r')
+plt.xlabel('time s')
+plt.ylabel('distance m')
+plt.title('cHPI w.r.t mean recording head position ')
+plt.show(block=False)
+
+###############################################################################
+# Plot raw data with annotated movement
+# ------------------------------------------------------------------
+raw.set_annotations(annotation_movement)
+raw.plot(n_channels=100, duration=20)
+
+
+# After checking the annotated movement artifacts, calculate the new transform
+new_dev_head_t = compute_average_dev_head_t(raw, pos)
+raw.info['dev_head_t'] = new_dev_head_t
@AdoNunes <https://github.com/AdoNunes> I have some ideas for improving
this script, okay if I push to your branch? It would be faster than writing
a bunch of comments
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#7096?email_source=notifications&email_token=AEILW3W4JG6VAGFHCU52WD3RAB2XFA5CNFSM4JQFA22KYY3PNVWWK3TUL52HS4DFWFIHK3DMKJSXC5LFON2FEZLWNFSXPKTDN5WW2ZLOORPWSZGOCTLIY7Y#pullrequestreview-349604991>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/AEILW3WZFCWPIPGKDYU3T43RAB2XFANCNFSM4JQFA22A>
.
|
I thought it would be didactically illustrative to show the original head position compared to the averaged. Let's see what Travis complains about...
 agramfort
left a comment
agramfort
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
|
Great!!!! |
|
it will be much faster for your next one I am sure :)
… |
|
Thanks @AdoNunes ! |
* artifact detection * art def * Rename file * style corrections * cropped * example cropped * flake corrections * added docstrings * fixed doc style errors * updatex example * fix docstring * added offset removal * added compute_average_dev_head_t * New head pos trans and annotate movement * Copy raw instance, corrected pos time * docstring fix * movement artifact only * Updated func names and example * enh doc & quat average * define norm * docstring correction * revive CI * quat 3 * docstrings improvement * logger and docstrings * docstrings * docstring, reference, latest * funct import * add func in init * import typo * quoted BAD_ * docstrings * FIX: Nesting test * added test * read raw * test def * docstring * docstring * test function * WIP: Tweaks * orig head pos line I thought it would be didactically illustrative to show the original head position compared to the averaged. Let's see what Travis complains about... * docstrings * warning to logger Co-authored-by: Eric Larson <larson.eric.d@gmail.com>
* artifact detection * art def * Rename file * style corrections * cropped * example cropped * flake corrections * added docstrings * fixed doc style errors * updatex example * fix docstring * added offset removal * added compute_average_dev_head_t * New head pos trans and annotate movement * Copy raw instance, corrected pos time * docstring fix * movement artifact only * Updated func names and example * enh doc & quat average * define norm * docstring correction * revive CI * quat 3 * docstrings improvement * logger and docstrings * docstrings * docstring, reference, latest * funct import * add func in init * import typo * quoted BAD_ * docstrings * FIX: Nesting test * added test * read raw * test def * docstring * docstring * test function * WIP: Tweaks * orig head pos line I thought it would be didactically illustrative to show the original head position compared to the averaged. Let's see what Travis complains about... * docstrings * warning to logger Co-authored-by: Eric Larson <larson.eric.d@gmail.com>
Closes #7077
I have added (temporally in the wrong place) a function script to mark suspicious channels of being bad, a muscle artifact detection (in practice almost the same as if Fieltrip) and movement annotation and head position trans correction.
The muscle and movement might be crucial in some datasets when Maxfilter is not applied and currently mne-python does not provide these functions.