-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
MRG, ENH: Add support for Hitachi fNIRS #9391
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
I'm not 100% sure on the answer to your questions, but we can try and puzzle it out together.
Could Wave be the equivalent of SNIRF-ProbeWavelength and WaveLength be the equivalent of SNIRF-WavelengthActual. If so, I would suggest we use Wave (the rounded values) in the channel name field, and store the precise wavelength in the info field (and use the precise value for the beer lambert calculation (currently we assume same wavelength for each pair, but would only be a line or two change to support different wavelength for each channel - although i think the affect of this on output would be negligible and other toolboxes don’t do it)). Location for precise wavelength could be something like mne-python/mne/io/nirx/nirx.py Line 311 in 3149819
It's entirely possible that these are not stored in the files you were provided, and are stored separately or expected to be known. For example, some vendors have just a few preset montages, if this is the case we could store a few preset montages (like we did with the Artinis devices #9141). Or, it could be that the vendor allows the optodes to be placed in any position, and does not provide a standard way to store the locations and leaves that to the user. In which case we could ask users to write an elc file. But this is all speculation, do you have a copy of the vendor manual or can you ask your collaborator how they unusually specify optode positions? I think you are correct that point 2 is same as 3.
No idea, but I think it many be bads like you suggest. Is it the same in all files? Does the vendor provide a standard set of files on their website? Just to ensure you actually were given a complete recording dataset (sometimes collaborators forget to send you parts of a recording and maybe they left out the montage file). |
|
I also have some Hitachi files I could test locally with. |
|
Okay I think it works testing locally with: My ordering of the If I use the Once I have the correct order in the docstring example this should be good to go, so feel free to test on your data. |
|
Sorry its taken me a few days to get to this @larsoner. I have run some of my data through your reader, and unfortunately it is not reading the data. Your reader works with Hitachi files with file version As these are just text files with a custom header I will post some file info I have below. I can also share some data with you privately if that helps? |
|
Here is an example file (with me having anonymised various aspects of the metadata)... |
|
@rob-luke I fixed some bugs and made the code more general. There is a bit of a problem, though, in that the nirs-toolbox code for getting the src-det pairs produces a result for example for 3x5: That both seems wrong -- these I think/assume are ordered the same way as the channels, but they have the first two channels with the same frequency for source and detector -- and does not match with the pairing in Hitachi 2 Homer: So I'm hesitant to use this to get your 3x11 pairings, and 3x11 is missing from Hitachi 2 Homer. Any idea where we can get the pairings? |
|
@rob-luke I think I figured out the pattern that gave rise to the Hitachi 2 Homer pairings, and I coded it and test it. Assuming it's correct and the iteration on the 3x11 is the same as the 3x5 (and not, say, column-wise instead of row-wise) then this should work. |
|
Even with the modification above I am still getting an error with my files. I will email you a file to test with. the error I am getting is |
|
Okay looks like it was a line-endings problem, should be fixed now @rob-luke |
 rob-luke
left a comment
rob-luke
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I tested this on a few local files and they read and return the correct data type and metadata. But I don't have access to a machine myself, so cant do detailed testing. From what I can test it looks good to me.
|
A collaborator had details of the electrode montage and what I get from MNE now matches their output, so I think we're good to merge |
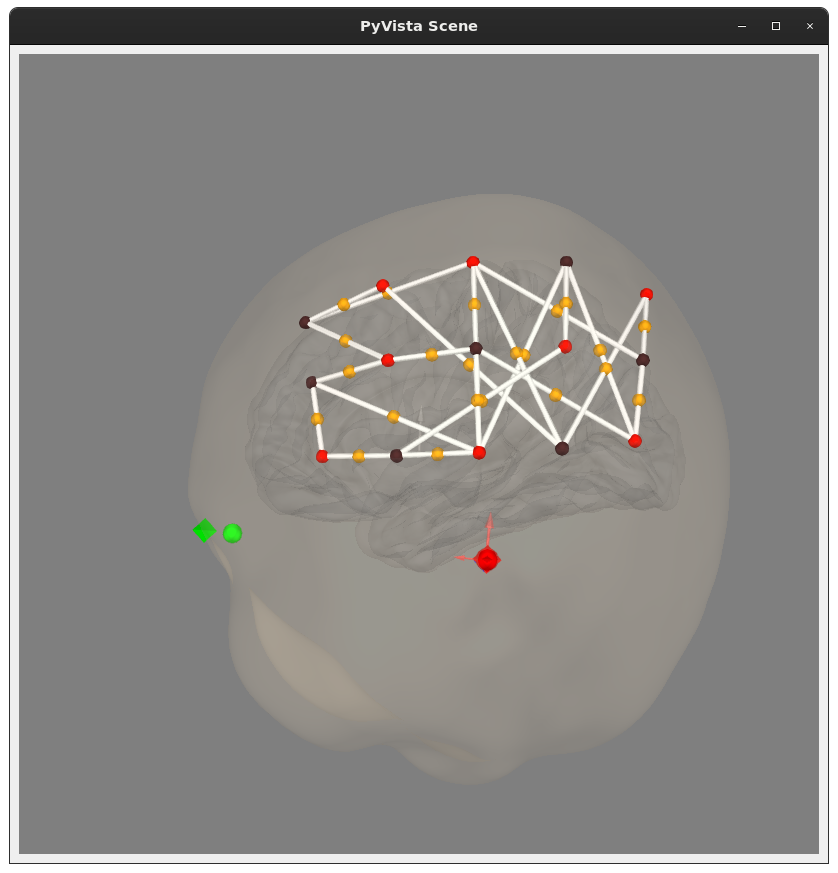

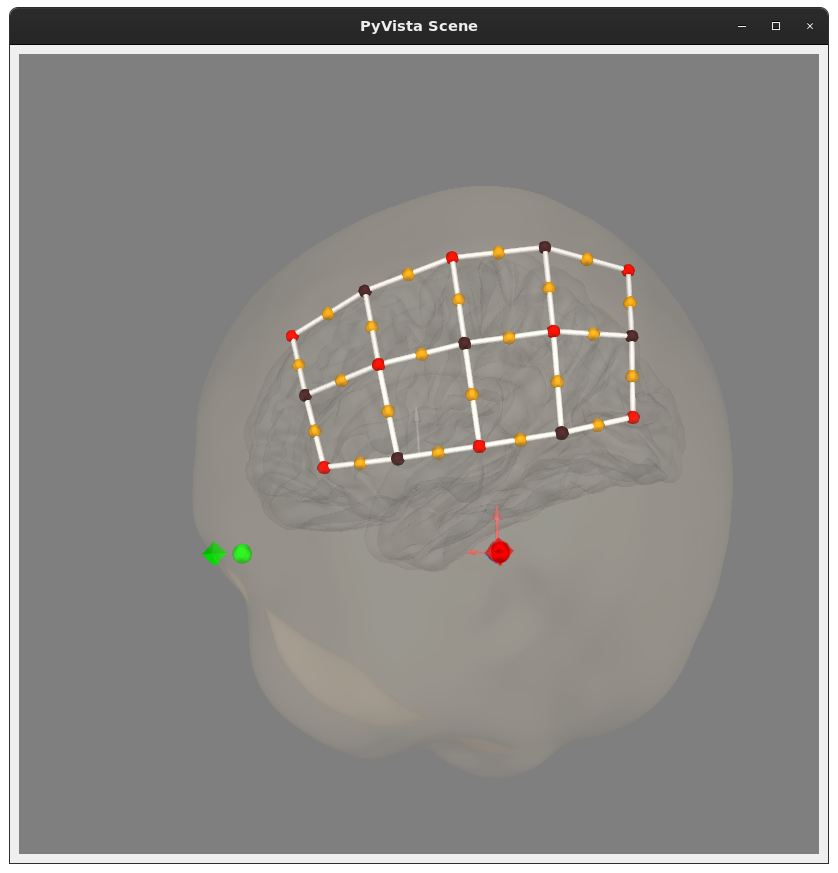
@rob-luke some issues so far:
Wave[nm],695,830in the header, but alsoWave Length,CH1(700.1),CH1(829.1),CH2(699.3),CH2(827.9),.... I forced the 695-ish ones to be 695, and 830-ish ones to be 830. Is this the right thing to do?S1_D1,S2_D2, etc. I thought about trying to construct this based on the wavelengths in the channel names, but there was at least one pairing that was identical in my file so that seemed like a no-go.Exception Ch,0,0,0,0,0,0,1,0,0,1,0,0,1,0,0,0,0,0,0,0,0,0means (should end up ininfo['bads'], maybe?)I can bother the folks who got me these data to get some of this information, but figured I'd check with you to see if you knew already.
Closes #9355