-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
MRG, ENH: Refactor MRI/CT code #9503
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
Oh wow |
|
Just pushed a commit to make CircleCI complete without error (my hack workaround for having none at the right value was wrong), feel free to push right over it @alexrockhill if you have a correct fix. |
|
Hmmm, yeah I just tried and it doesn't work either. I think that's because the rigid registration can't be zoomed... Do you want to split it up into two steps? |
|
The reason |
You must mean for the CT-MR warp step in the subject's own space. I'll I add a
Is this still in reference to the CT-MR warp, or the MR-template warp? |
In reference to the MR-CT registration which I don't think qualifies as a warp and so I think is misleading. |
|
I think it should be split into two separate registration and warp steps, I can take a pass if you'd like |
To me it's all registration/warping/coreg, just with different levels of granularity (translation is one, rotation is another, affine-but-still-linear / shear / scaling is another, SDR is another). This is why I'm inclined toward a single function to do it... |
|
I made the first step rigid and it still has invalid electrodes |
If it has the same |
I just think when you're passing a CT into a function to align with an MR, you don't want it to be "warped", that would be bad, just registered from my best guess at a naive user's perspective |
|
Maybe a good solution is to call it That's just my perspective though, maybe I'm being unreasonable and others are perfectly fine having rigid registrations under morph and warp. |
|
Maybe under |
|
... I think you're right that |
|
Okay all electrodes get assigned now, feel free to look at the outputs to see if they make sense, and also the naming/structure etc. |
|
Looks great! I don't have write access but I would suggest having |
This suggests we might actually want the entire set |
|
I think also a keyword |
How about |
That sounds great except that I wouldn't want it to carry on at a lower resolution. If the previous level used some downsampling, that doesn't mean the next level has to, in fact in the case of the CT to MR, you want to zoom and then not. On |
I think that's getting really confusing to me. Maybe a In Should be a tuple if the default is |
That seems like a nice way to do it!
So the logic would be "native unless specified"? I can live with this, especially if we alias |
Sounds perfect, totally on the same page |
|
Okay I'll get to it tomorrow, or if you want to give it a shot you can |
|
I didn't finish and I need to fix the commit history but I pushed these in case you want to finish it tomorrow morning before I'm up, but I can also fix and finish tomorrow just a bit later, I'm on PST. |
mne/transforms.py
Outdated
|
|
||
|
|
||
| @verbose | ||
| def compute_volume_registration(moving, static, steps='warp', zooms=None, |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I would call moving_img and static_img to suggest it's a nibabel Image
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I had it that way at first but thought moving and static alone were clearer because it matches what dipy calls it in affine_registration:
https://dipy.org/documentation/1.4.1./reference/dipy.align/#affine-registration
So here I think being consistent with their naming is worth it. (Plus the name is shorter.) But I don't feel strongly so if you aren't convinced by the consistency argument I'll change it
This reverts commit 6cd2500.
This reverts commit 22f3f14.
This reverts commit 07c07e3.
This reverts commit 288cd91.
This reverts commit 97c43cd.
This reverts commit be9154a.
This reverts commit cb132a6.
fix core fix affine map working version for tutorial, not tests fix test
This reverts commit 50e8107.
|
Okay @alexrockhill feel free to have a look and see if you're happy with the code, here is the updated example: https://30168-1301584-gh.circle-artifacts.com/0/dev/auto_tutorials/clinical/10_ieeg_localize.html |
|
Windows is having a bad day because of the VTK 9.0.2 release, can be ignored here as I'm working on it in #9512 |
Still looks off... I would rather have a maximally accurate example that has a lot of code that's not abstracted than one that doesn't work quite right. I think the existing morph functions may not be compatible with the what is on main now for the tutorial. I think first priority if you want to go this route of refactoring is getting a version that works and then maybe in a followup we can get the SDR morph from the volume source estimate working using the same code. I really would rather not spend a ton of time debugging the volume source estimate code so I think it's best to part ways. I'll take a pass now. |
Can you clarify how/where? I'm assuming you're talking about subject->fsaverage and not CT-MRI. For "to fsaverage", those temporal electrodes are now nicely on/around the actual lobe, which is what I was always worried about, so to me it looks okay. Is there an image above somewhere (or in some artifact) that looks better?
Can you see if just
So maybe as an intermediate, if zooms=None does not work, we can keep the MR-CT alignment that is here currently since it works, then put back your custom dipy code for the subject->fsaverage alignment and save those warped electrode and brain images? |
The CT-MR is way off still, that's the main thing. The SDR looks pretty good. |
So this is what we have now: And this is what we have in I think it only differs by 1.2mm and 1.9 degrees from what you had before. I have a hard time seeing meaningful differences here, and if I didn't know which is which I'm not sure which I'd say was better, but I am no expert here! As much as I'm inclined to keep it, indeed we shouldn't keep something if you think it is "way off" even if I can't see it. I thought you had said before that the MR-CT was now okay, but since it's not I agree perhaps the easiest thing to do is just to revert to the old affine and commented out code for the MR-CT step, and revisit sometime later how to use our standard pipeline to get an affine of equivalent quality. |
Way off is a relative term and I think you have to spend some time aligning these by hand to appreciate when the fit is right and when it's not. 1.2 mm and 1.9 degrees isn't a whole lot but those are clearly how much it's wrong and to me, that's too much. You can align by eye closer than that, the whole point of this is that it should be as precise as possible with the alignment, better than what can be done by human eye. I could fix the top image to be much closer by hand. |
|
Keep in mind that you're only seeing a bit of the alignment with the three slices and in a viewer like freeview it would be much more obvious how much the alignment is actually off and I would consider the first alignment to be poor and need fixing if someone had done it by hand and I was reviewing it and the second alignment would be good. |
| alignment_affine = np.array([ | ||
| [0.99235816, -0.03412124, 0.11857915, -133.22262329], | ||
| [0.04601133, 0.99402046, -0.09902669, -97.64542095], | ||
| [-0.11449119, 0.10372593, 0.98799428, -84.39915646], |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@alexrockhill okay if I just revert the text removal here, then we're good to merge?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
That's the correct alignment but the commented out code should return that matrix and it doesn't right?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
By revert the text I mean both this affine and the old commented code, which I assume is what you used to generate it
|
Ok I just pushed a different pull request to split this up. First, I think a version that works with the tutorial should be merged which is the PR I just pushed. Then we can try and refactor the volume source estimate code to use the new transforms. I think combining the two steps is causing too much of an issue. |
|
Closing for #9515 |
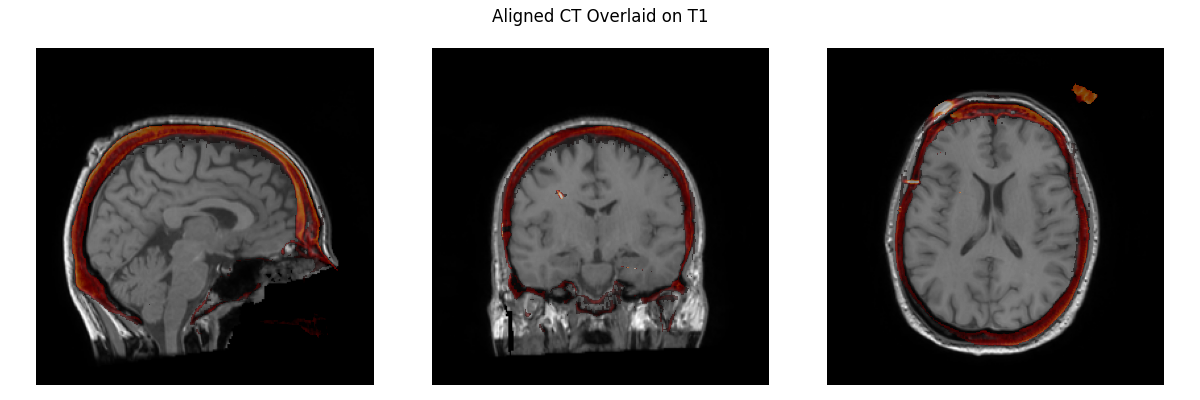
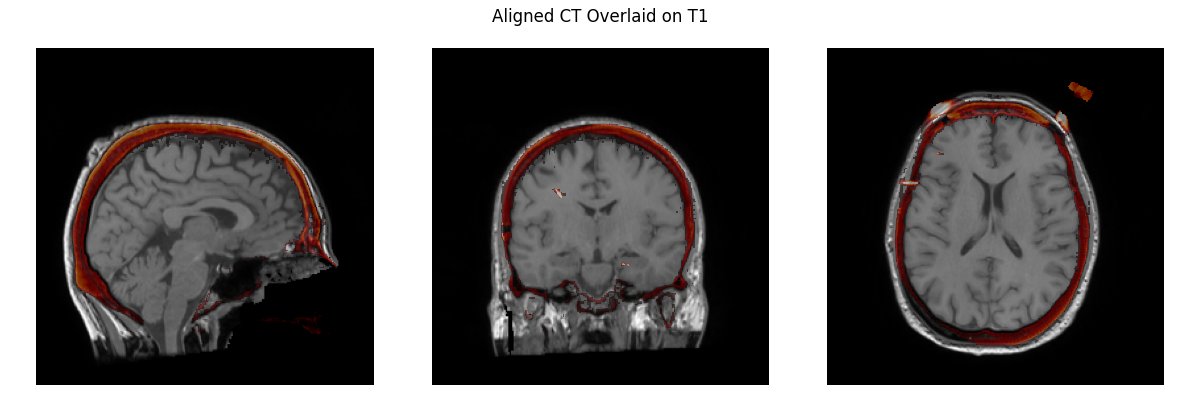
@alexrockhill can you take over the first two steps below:
np.wherecan be length zero, which is bad)Then I can do for the functions I added:
71.64 sec 2537.8 MBSourceMorphobject attributesniter_*->niterdict, addpipeline