-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
[ENH, MRG] Add mne.viz.Brain.add_volume_labels #9540
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
Ok this is missing all the brain stuff but I was a bit too tired to figure it out tonight. Looks like it works as a minimal version though. I'll add some tests tomorrow or maybe I could get a little help finishing this one because the |
|
Using: I get: So I added some smoothing and now I get: I'll push, then @alexrockhill I think if you add this to |
|
Awesome! Looks great!! Sure, I can do the last bit when you push your version. |
|
I'm done, feel free to push! |
|
Wow you put labels on each of the default lut indices, thanks! |
|
Yeah it was a quick copy-paste-trim from the FreeSurfer LUT file. vscode makes it really easy to edit N lines in an identical way :) |
mne.viz.Brain.add_aseg to plot anatomical segmentation surfaces
|
Hmmm the electrodes seem to be out-of-scale as well... it might be easier to mock up a quick This is with |
|
You need units='m' in the brain constructor |
|
(plot_alignment only ever uses meters but brain defaults to millimeters for consistency with freesurfer and pysurfer) |
mne.viz.Brain.add_aseg to plot anatomical segmentation surfacesmne.viz.Brain.add_aseg to plot anatomical segmentation surfaces
|
Last run CircleCI complained: Not sure if you fixed those in the latest rebase or not... |
That's going to depend on #9545 to merge first but other than that, yes taken care of |
|
@alexrockhill this needs a rebase or merge. I went to do it but I think you'll have ideas for how best to combine Also based on naming consistency I suppose this should be |
|
@alexrockhill I pushed a commit that cleans up the smoothing and makes the marching cubes faster for labeled volumes following the VTK example https://kitware.github.io/vtk-examples/site/Cxx/Medical/GenerateModelsFromLabels/ |
|
... so I think all we need here is a rebase or merge with upstream/main, and rename |
mne.viz.Brain.add_aseg to plot anatomical segmentation surfacesauthor Alex <aprockhill@mailbox.org> 1626322812 -0700 committer Alex <aprockhill@mailbox.org> 1626466592 -0700 add roi/subcortical plotting wip wip wip slighly working version FIX: Fix units and render ENH: Cerebellum wip add legend, fix seeg tut add legend kwargs, update visualize stc tut fix flake fix doc, fix copy error in tutorial fix func->meth Eric suggestions
|
... just realized you asked about |
In this case I would favor a PR to |
Yeah I can open an issue for that too, I was able to just change the view angles for now. |
If it's as easy as adding |
|
Interestingly, it ran locally for me with the insula label but that was for Actually that's for |
I suspect it's a problem with the example arguments/usage. Can you fix it? |
Sure in a meeting, will fix in 30 minutes |
Sounds good, FYI two examples appear broken now: |
|
I think some changes maybe to smoothing have broken it, it's not finding any labels not just insula. Looking into it |
The failure is in the |
I'm going to try putting things back before your latest marching cubes change (only the first smooth), if that fixes things then we can do the discrete part in another PR. |
Feel free, but please preserve the warning. I suspect things might have been failing silently before (because we just silently didn't bother trying to do marching cubes before my commits) and the warning makes it clear that something is wrong somewhere (we shouldn't in an example ask to plot a volume label that does not exist in the mgz) |
mne/viz/_brain/_brain.py
Outdated
| for label, color, (verts, triangles) in zip(labels, colors, surfs): | ||
| if len(verts) == 0: # not in aseg vals | ||
| warn(f'Value not found for label {repr(label)} in: ' | ||
| f'{aseg_fname}') |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is the warning we need to preserve
Good call, it does look like what was broken was from the marching cubes changes though. I didn't really delve into it fully but when I reverted all the surfaces rendered properly |
|
So there was an issue with |
Did you verify this by locally testing with some
So pushing commits that uniformly keep CIs green is better than pushing commits without checking locally that things work first. The CIs are really meant to act as a sanity / completeness check of things that have been checked locally.
Can the discrete support be restored, or is there also an issue with the smoothing there? |
|
Yeah sorry, got a bit hasty with the fix for the last PR being a priority. I checked and running locally there are no warnings for the
There was an issue with the atlas in the tests but also a larger issue with the discrete method, I don't think it was working... |
Indeed it failed to find some ROIs, if I force it to use VTK_DOUBLE it seems to work fine! But like you said I can open a separate PR for this once this one is merged. |
|
Not waiting for macOS on this one, thanks @alexrockhill ! |
|
Sounds great, looks like it's good to go |
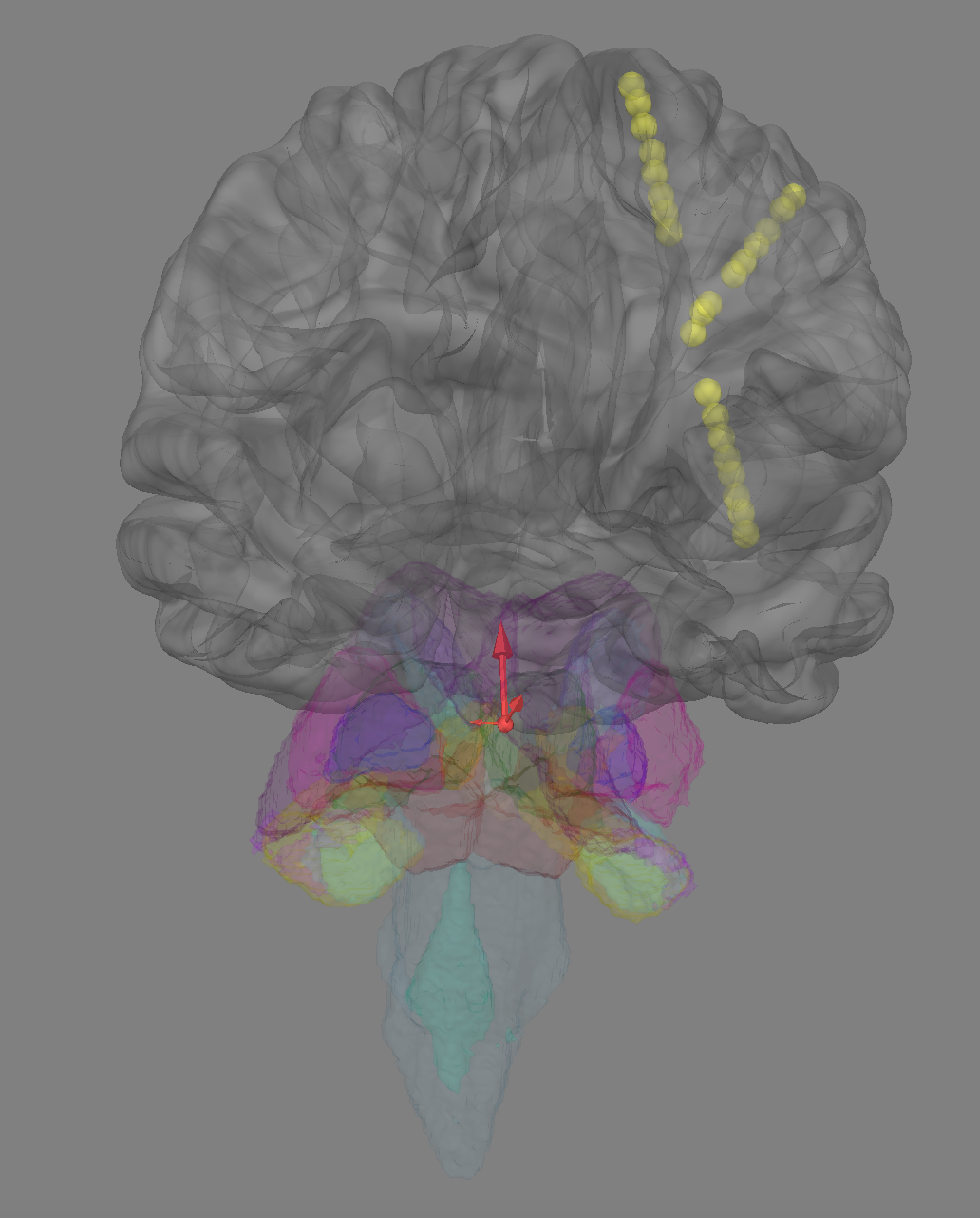
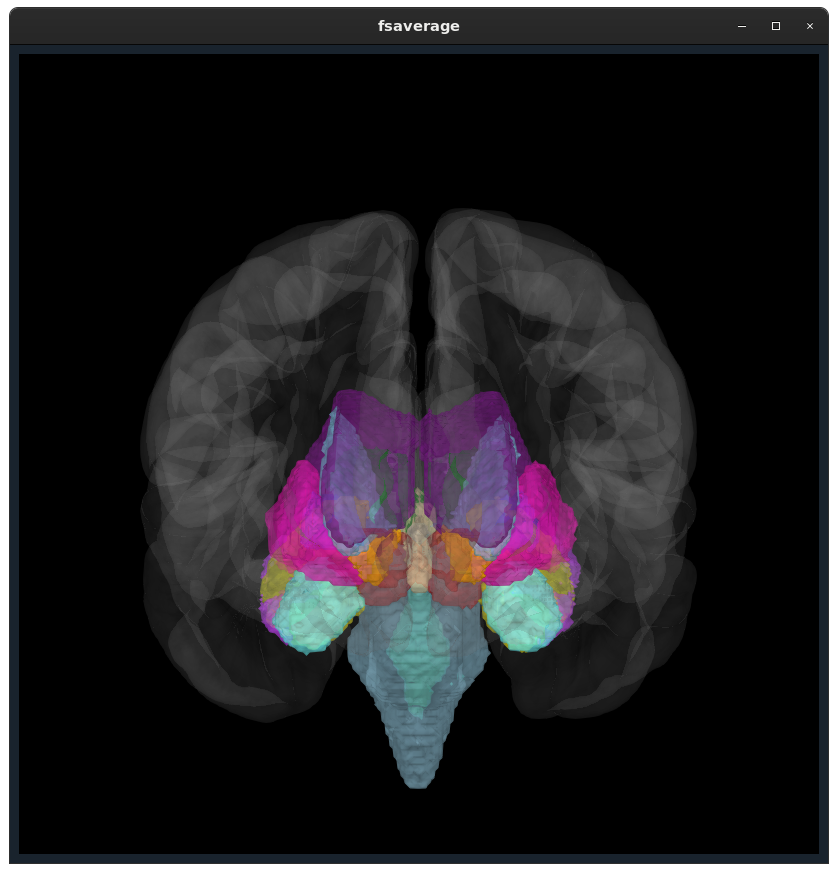
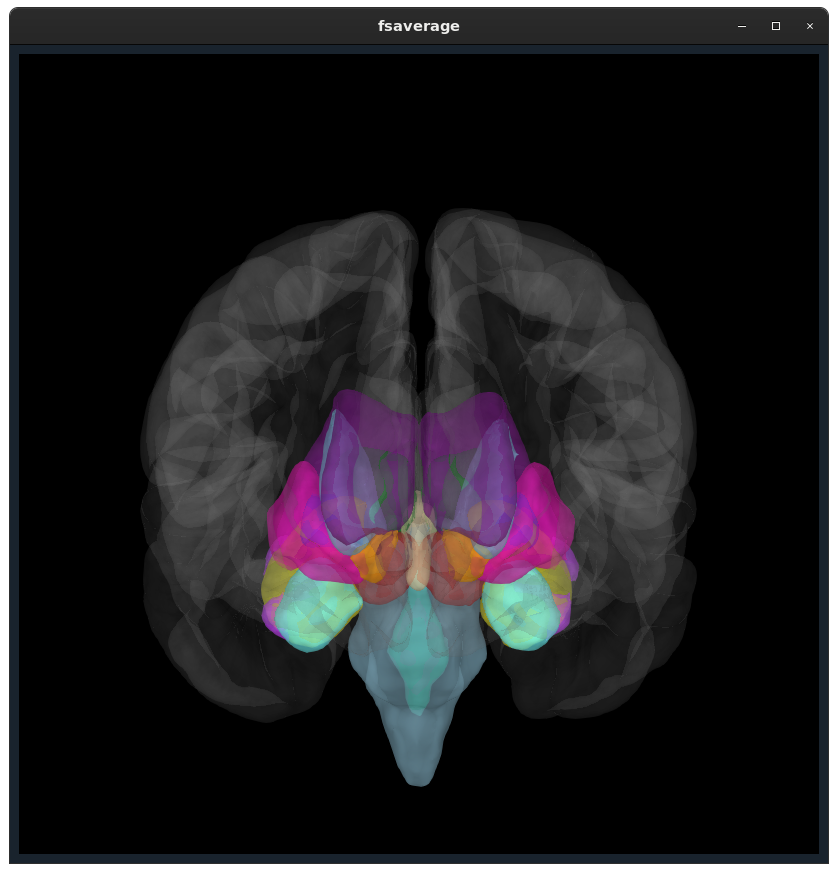
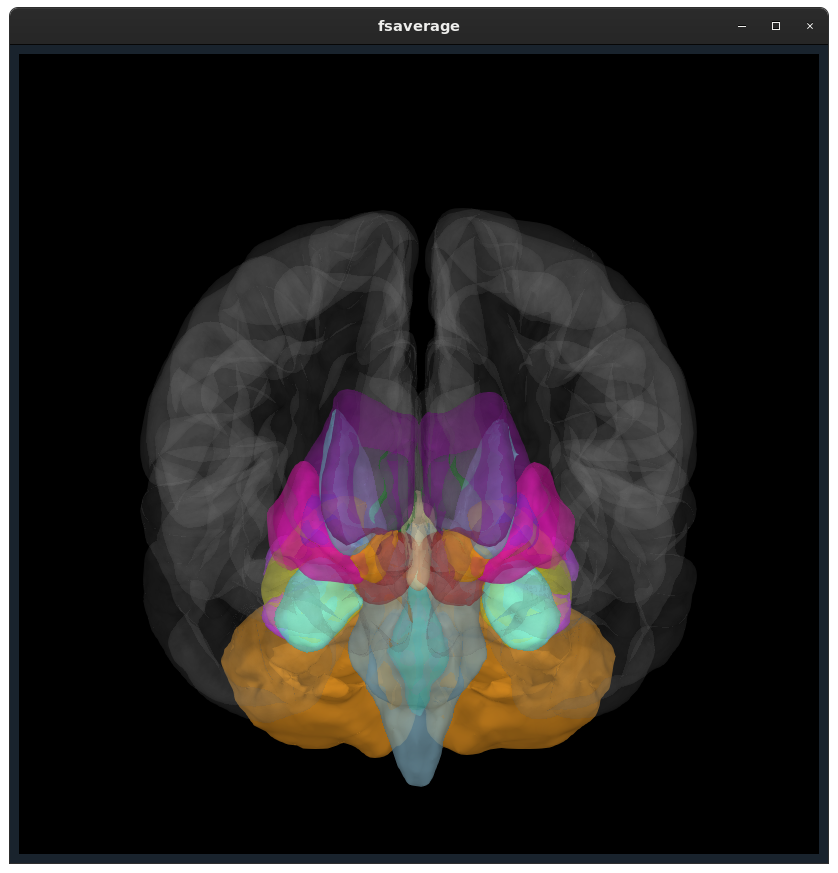
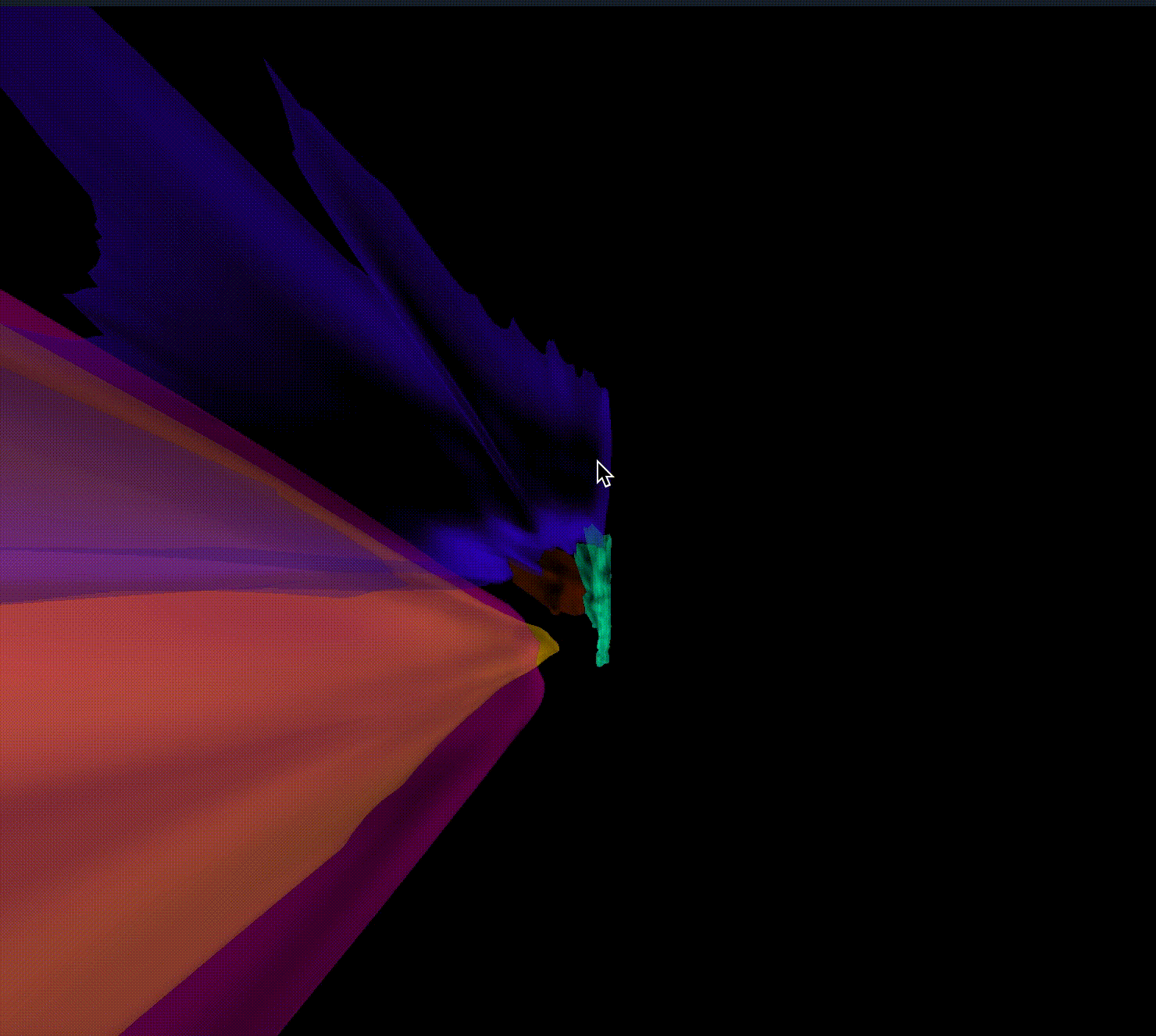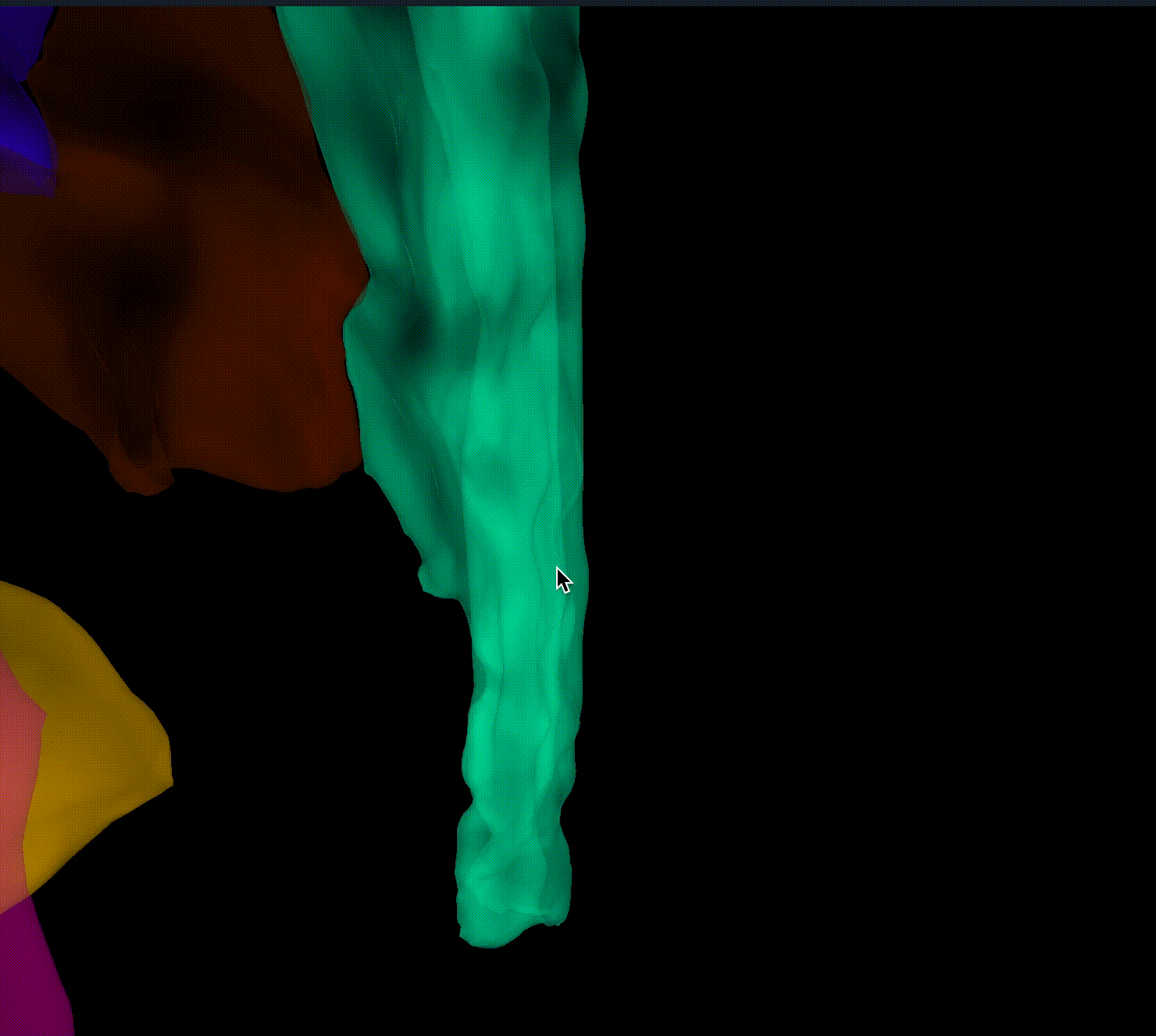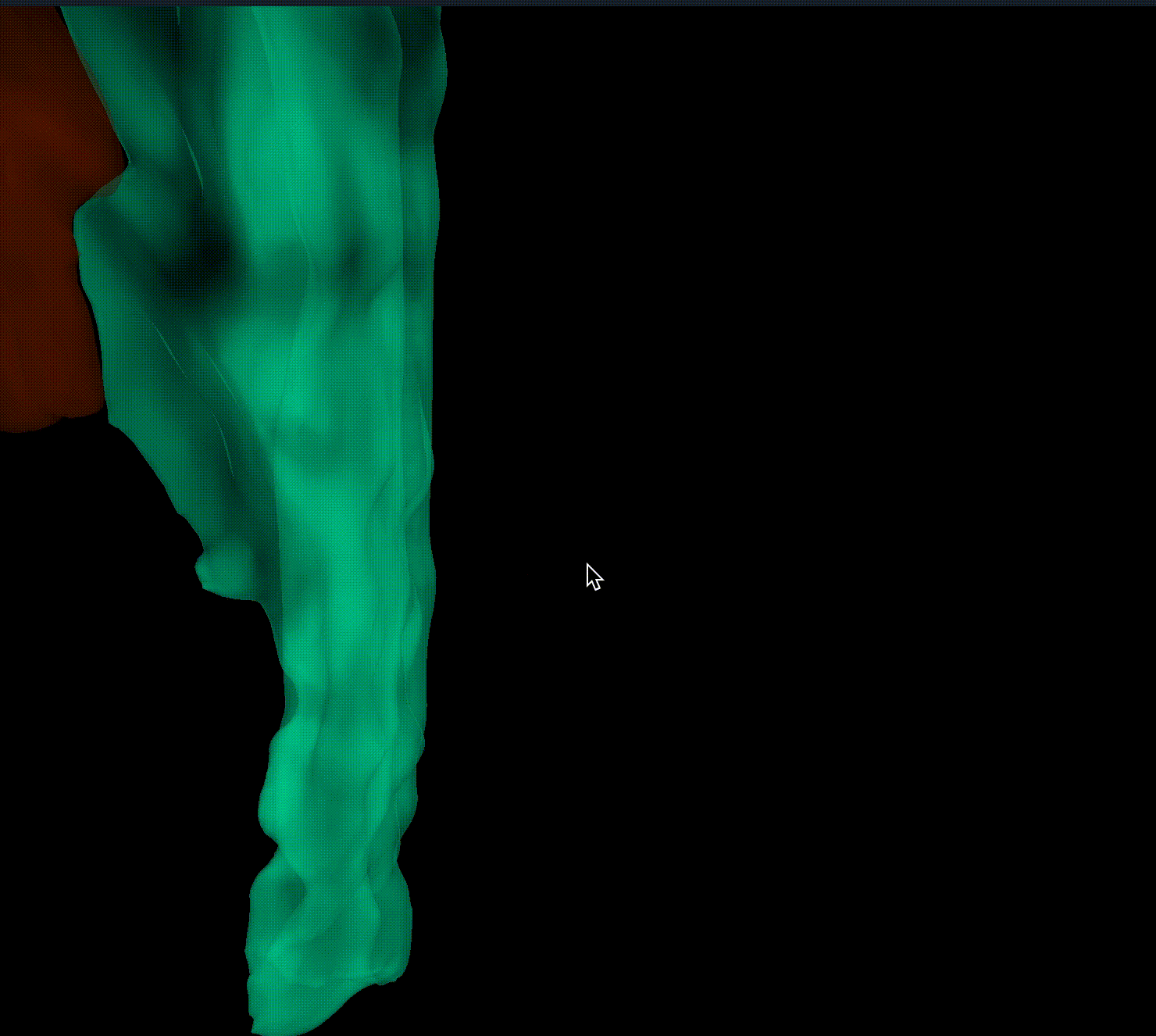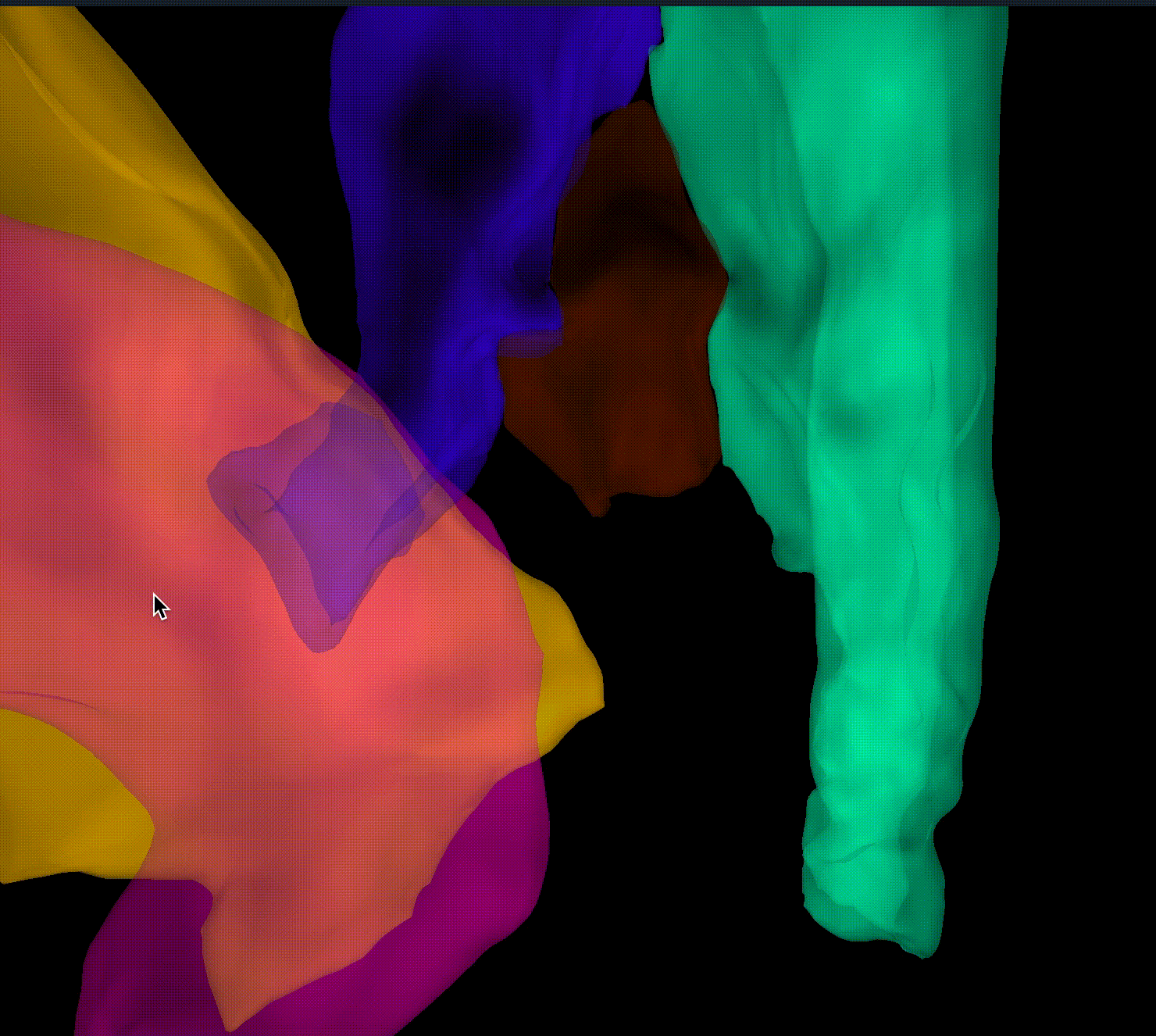

Closes #9520
Here is a very, very rough draft of what I was thinking, I didn't want to go too far in a bad direction so if you have a second to look at it @larsoner, that would be very much appreciated.
Basically, the ROIs are a very different kind of surfaces than the SUBJECTS_DIR/SUBJECT/surf surfaces and so I think need their own class but I think inheritance works nicely; they are named very differently and don't all have hemispheres.