-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
[ENH, MRG] Compute Electrode Bridging in EEG #10571
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
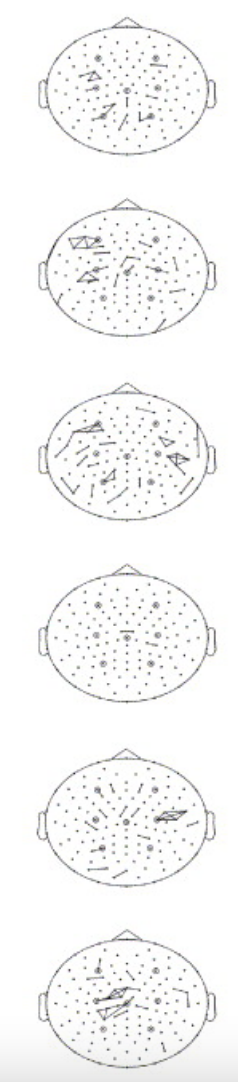
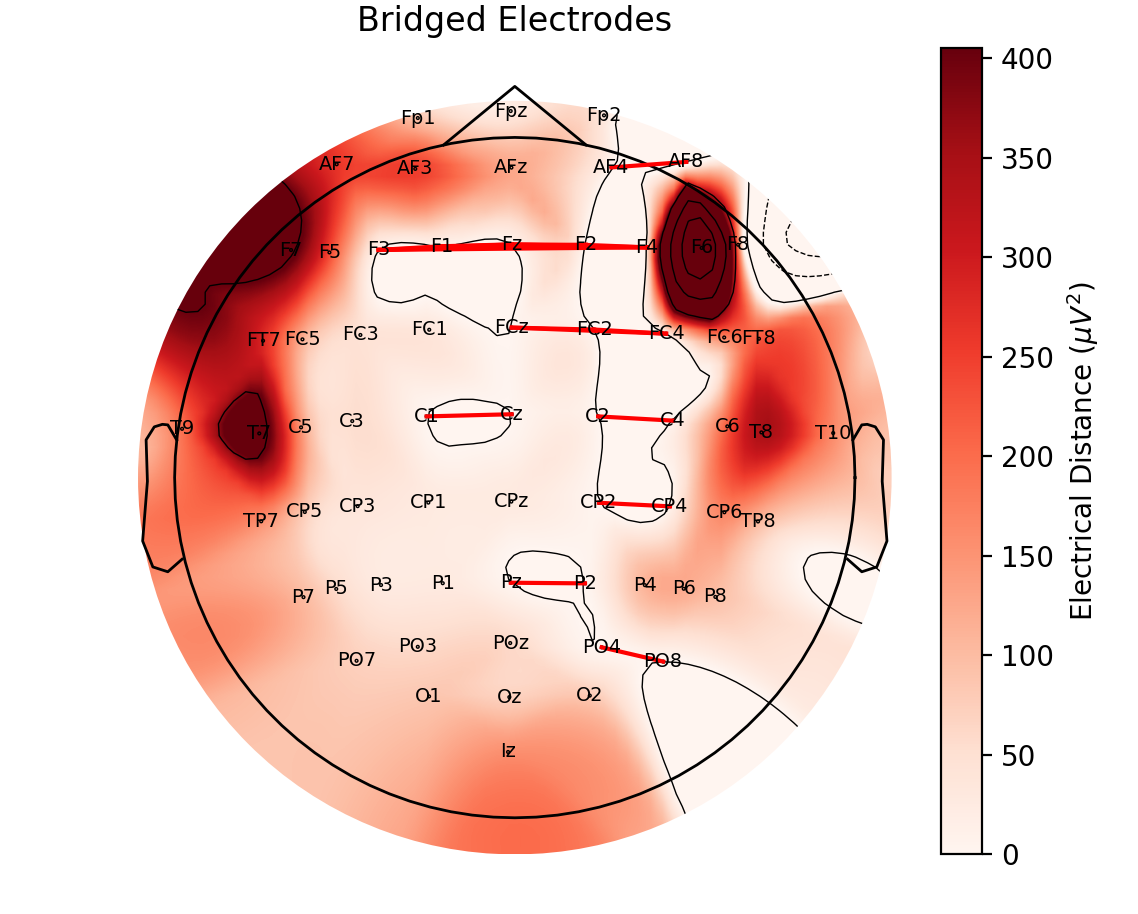
The second plot here https://output.circle-artifacts.com/output/job/01cda15a-cc39-48d4-acf5-133082e67e2b/artifacts/0/dev/auto_examples/preprocessing/eeg_bridging.html has a preponderance of lateral connections whereas these bridges seem more evenly distributed https://doi.org/10.1016/j.clinph.2003.10.028 |
|
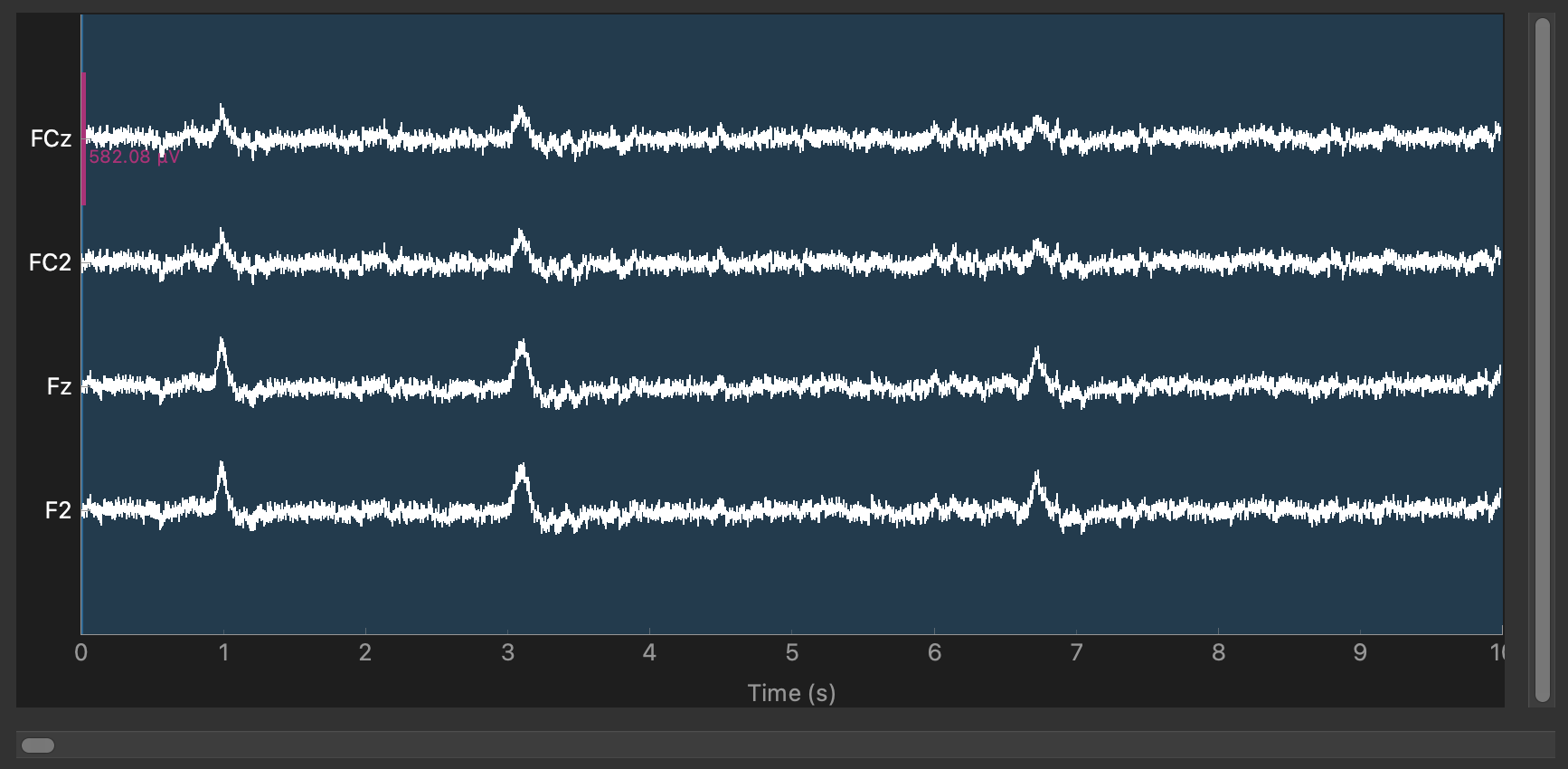
Ok, I'm starting to think this is just a legitimate pattern, maybe having to do with the EEG experimenter applying gel from the side and so it bridges laterally for this dataset. Here are two channels where the top two are bridged and the bottom two are bridged but they are not all inter-bridged:
If anyone wants to look this over/comment, please do, but I assume this is pretty much on track from this evidence. |
|
Ok this ended up being a lot more work than I thought but I'm really happy with the result.
The only question I have currently is that it's a bit long as an example so it could be moved to the tutorial section. I really like the deep dive because I think this is an important topic in EEG and is doable to understand in a way that will help people collect data better. |
|
Thanks for the review, sorry for the typos 😊 Should pass the tests now. |
|
The test that timed out might need to be rerun but ready to merge by me Wait muV^2 still got messed up one sec |
|
Hey finally got the formatting right https://output.circle-artifacts.com/output/job/482745b0-9c03-4112-be95-892f9f3f97a7/artifacts/0/dev/auto_examples/preprocessing/eeg_bridging.html, okay looks good. |
|
@cbrnr or @mmagnuski would you have time to read the example https://output.circle-artifacts.com/output/job/482745b0-9c03-4112-be95-892f9f3f97a7/artifacts/0/dev/auto_examples/preprocessing/eeg_bridging.html and see if you find it appropriate? |
 agramfort
left a comment
agramfort
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'd love to have another review from one of our EEG experts
|
I'll take a look. :) |
 mmagnuski
left a comment
mmagnuski
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is a great tutorial! I never actually checked for bridging in my data and now I'm eager to do it. I think it would be useful to add some recommendations in the summary towards the end:
- for example to record a few minutes before starting the experiment
and have a script ready that plots bridges / warns about their presence? I'm just not sure what can be done if channel bridges are detected at this point. - what would be recommended course of action once brides are detected during preprocessing? Dropping or interpolating some of the channels if there is not too many brides? I don't know as I never actually checked for this problem (regrettably).
Something real-time would be very cool but once the bridges are created, to my knowledge there is nothing you can do about it. So I think the only feedback you would get in real time would be whether to abandon the session or not, which I don't think is really all that helpful. I think the better use would be post-hoc like this and to just see how many you bridged and try to do better in subsequent experiments. You can interpolate the bridged channels for sure but that's at least two adjacent channels being interpolated... |
|
Does this look okay now @mmagnuski? |
|
Yes, looks good! |
|
oh, I forgot one thing: Thanks @alexrockhill ! |
Fixes #10561.
This tutorial is pretty compelling even though there are a lot of parameters https://psychophysiology.cpmc.columbia.edu/software/eBridge/tutorial.html so I just did an exact replication despite there being a few different strategies in the literature that depend less on the histogram of electrical distance values and more on the projection of the nearest neighbors onto each electrode (intrinsic Hjorth) e.g. below.
However, I'm getting a weird effect where the bridged electrodes are predominantly lateral to each other in the EEGBCI dataset. If anyone had a second to look through the code, it's not that complex but I'm guessing I did something wrong.
I'll keep looking as well and hopefully fix it soon. I think this is pretty valuable for EEG users.