-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
MRG: Automatically selects EEG channels when plotting bridged electrodes on topomap #10753
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
I don't think the failures are related to this PR. |
 agramfort
left a comment
agramfort
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
 alexrockhill
left a comment
alexrockhill
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Looks good, one small nitpick.
| topomap_args.setdefault('image_interp', 'nearest') | ||
| topomap_args.setdefault('cmap', 'summer_r') | ||
| topomap_args.setdefault('names', info.ch_names) | ||
| topomap_args.setdefault('names', pick_info(info, picks).ch_names) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Nitpick but you call pick_info twice, would be nice just to do once.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, I thought the same, but I did not see a clean way to pick only once (at least not without changing more extensively the function). Problem is:
- you need the 'picked' info at the beginning to set the 'names' argument (alternatively
[ch for k, ch in enumerate(info.ch_names) if k in picks].. not super clean and extends a lot this line). - you need the 'picked' info when plotting the topomap with
plot_topomap - you need the 'non-picked' info at the end when adding the bridged connections because
bridged_idxis based on the original info with all channels.
And the gain with the list comprehension for the 'name' argument is only ≈1 ms but loses in readability.
Feel free to add further changes if you have a clever way to pick only once while managing the bridged_idx, I didn't spend much time searching for one ;)
|
Also @alexrockhill, another bug I want to address in a different PR (probably tomorrow as I'm getting started on this electrode bridging preprocessing step that was missing in my pipelines) -> If you run the snippet below on main or if you run the code snippet in the first post that raises on main and is fixed in this PR (i.e. I did not break this 😅): Outputs: The line doesn't look like it's at the correct place ;) EDIT: I actually dig a bit instead of shutting down the PC.. it has to do with the sphere radius that is off and not equal to the default The code snippet above does force the sphere radius to the default value and does produce the correct figure. Without this, the sphere is set to It looks like it's a combination of 2 bugs:
|
|
@mscheltienne I'm not sure about the second bug, I'll have to look into it. Can we just use the |
mne/viz/topomap.py
Outdated
| See also :func:`mne.preprocessing.compute_bridged_electrodes` to determine | ||
| ``bridged_idx`` and ``ed_matrix``. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The function name should go into a See Also section
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
For the See Also section, it looked to me like usually it only contains function x-links without any text. Can you also put 'comment' like this one which mentions 2 of the arguments in the See Also section?
|
@alexrockhill @hoechenberger The example runs correctly locally and the figures are correct. One of you can can merge if you don't have further comments :) |
|
+1 for MRG on my side when CIs are green |
|
I pushed a fix here for the sphere problem. The issue is: Notice the additional info that is now used to auto-fit a sphere in the second case.. |
|
As green as it will get with the hash issue on the testing dataset. |
 hoechenberger
left a comment
hoechenberger
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Pushed a small docstring change, otherwise LGTM!
|
yes, this is unrelated but disheartening, as I thought @larsoner and I eradicated that bug a week or two ago :( I'll look into it. |
|
Thanks @mscheltienne for fixing this, I really appreciate it! |
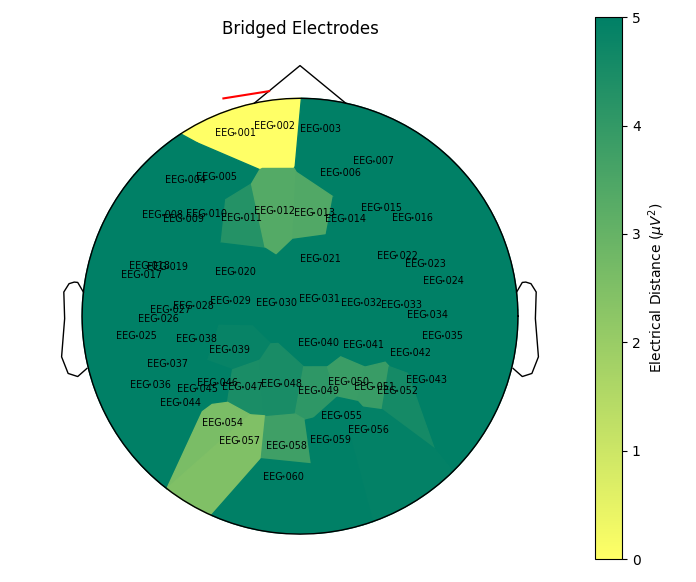
I was playing a bit with the new bridged electrode detection feature introduced in #10571 and I ran into a couple of issues with the plot on topographic maps.
As for
compute_bridged_electrodes,plot_bridged_electrodesshould automatically selects the EEG channels. There was an attempt here:mne-python/mne/viz/topomap.py
Line 2762 in 5944b87
infowhen creating the topographic map.Snippet that raises on main: