-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
[ENH, MRG] Add sensor projection for ECoG #9800
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
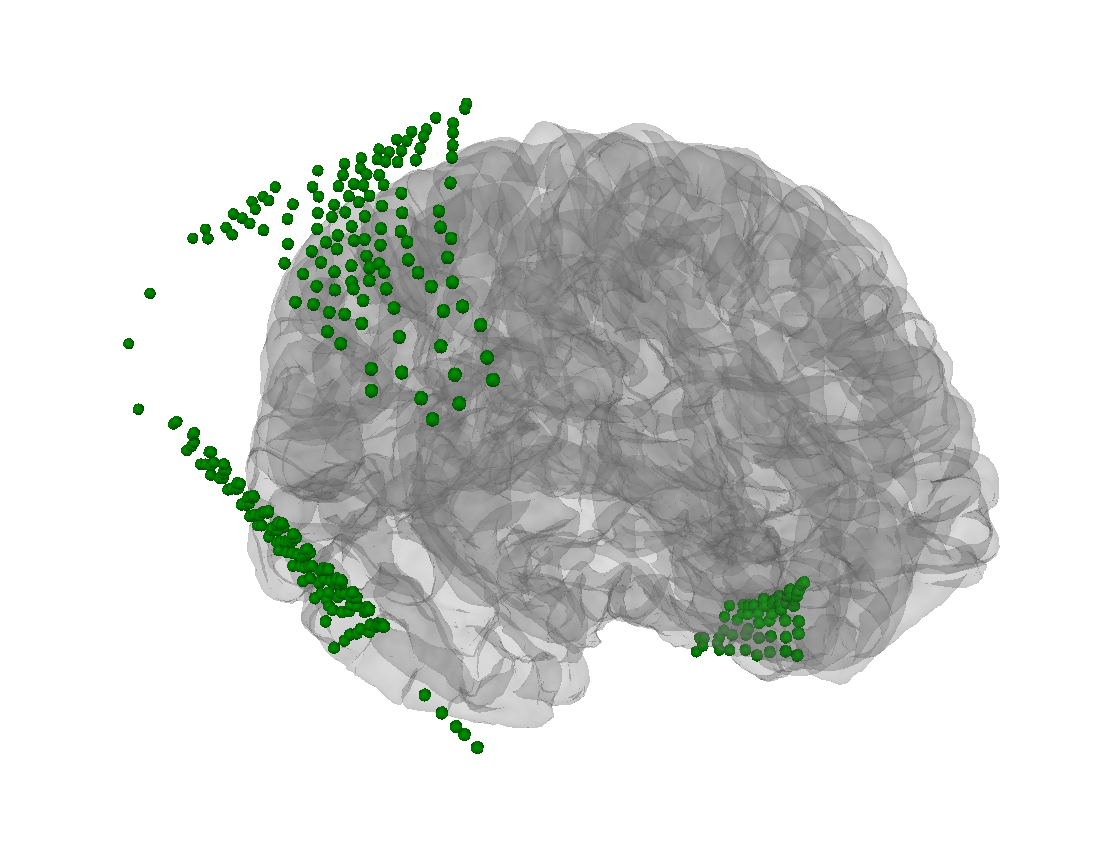
This looks okay to me besides that there is a split across the gyrus in the larger array https://35145-1301584-gh.circle-artifacts.com/0/dev/auto_tutorials/clinical/10_ieeg_localize.html Any ideas on how to fix that? |
 larsoner
left a comment
larsoner
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
My only idea on the gyral jump is that the brain surface might have some bump/irregularity causing a problem. Have you plotted by eye to see if it makes sense? Some plot_alignment call with surfaces=dict(brain=1.0) should make it clear I think. If it's wrong, sometimes you can fix it by tweaking the FreeSurfer calls in the right places...
If we can't fix it, maybe just make a note that it's probably incorrect for these data but it's the best that we can do? Keep in mind that we sort of know FreeSurfer doesn't make perfect BEM surfaces at this point (though they're usually "good enough" for M/EEG), see for example #8840, #8825, and a host of other similar issues over the years.
From an API standpoint I wonder if this should live in a namespace like:
mne.preprocessing.ieeg.project_sensors_onto_brain
similar to how we have fNIRS-specific stuff in
mne.preprocessing.nirs.beer_lambert_law
It helps make it clear that this is really a data cleaning/fixing step, and that it's really only applicable to iEEG data. (At the end of the day we need to put less stuff in mne. rather than more when possible... it's already too crowded in there.)
It doesn't look like the brain surface is the issue...
I wrote it so that it could work for any sensors to any surface so that it would be more general in case there was a use case in the future. I think a refactoring of ieeg or intracranial-specific code including what I've added recently that uses marching cubes would be good but I think that would be better as a separate PR like the |
Except that this adds a new public API, so we should decide the right place for it to live before merge, rather than after |
Doesn't the brain surface come inside the pial surface in many places? That doesn't look right to me -- it should be strictly outside the |
|
Ooof especially that right temporal pole |
This is the very last public API in the intracranial steps, everything else is done, I just was focusing on SEEG but this is good to add for completeness. The last one seems like maybe not the best time for major refactoring... |
I'm not sure this is the source of the problem, I thought it was because the center of the grid was farther away from the surface than the edges leading to a bifurcation point when it's closer to one half than the other, not sure though |
I'm not really talking about any major refactoring regarding the public API, just where the newly added (in this PR) Separately, I mentioned that the internal implementation of Does these two ideas make sense? |
|
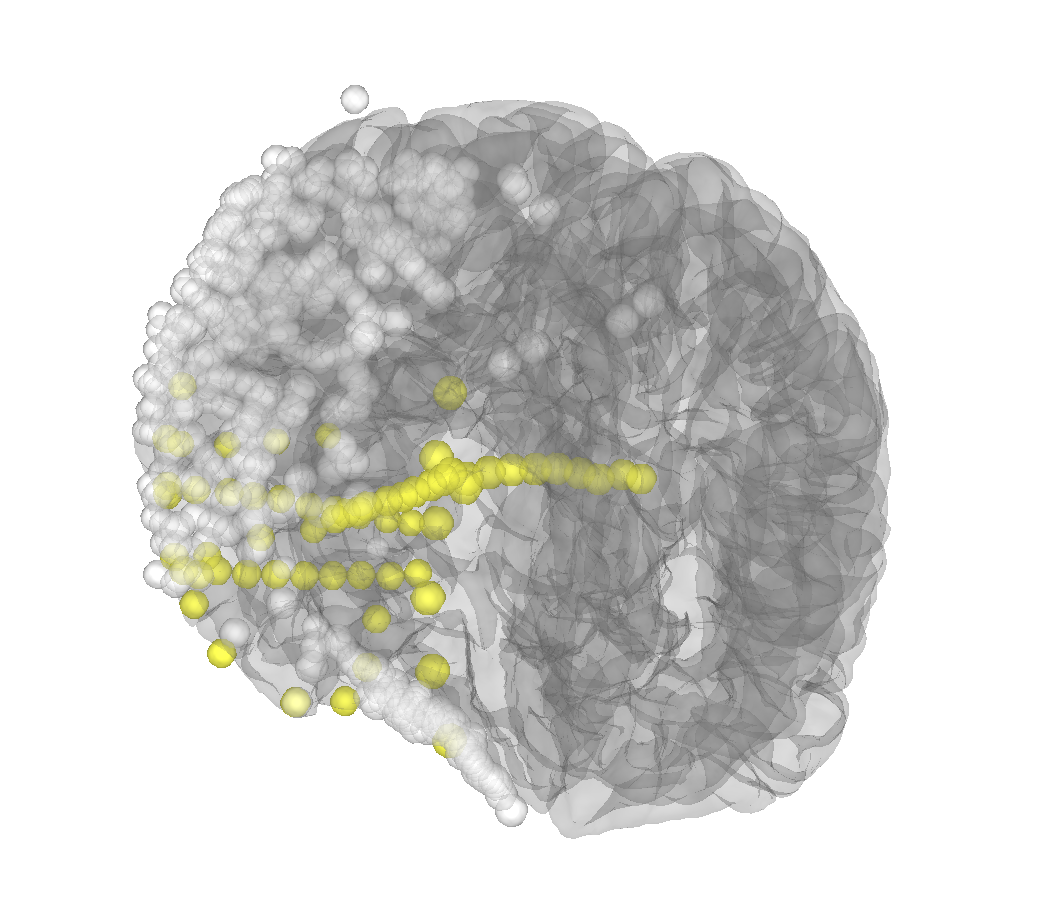
That does seem to be the issue (i.e. the brain surface is okay) because the projection onto inflated looks like this It looks like there are two papers cited by iELVis on how to deal with the issue but their implementation is a bit complex https://www.biorxiv.org/content/10.1101/069179v2.full... |
I'm not sure what you're plotting, except that the green dots are outside not just the skull but even the scalp (esp. for the parietal/occipital electrodes). To me this looks like a head<->MRI problem, but it could be something else... |
Okay you updated -- you cannot project onto inflated and expect it to make sense, its geometry should be thought of as (mostly) independent of that of the realistic pial/white/brain/skull/scalp surfaces. It basically shares a triangulation with pial/white, but the positions don't make sense. IIRC it even is centered at the origin, which clearly is nonsensical for a hemisphere. (There is some geometric gymnastics done in |
|
Okay well it seems like this simple projection doesn't really do the job and my best guess is because of the geometry. The way it has been solved before involves considering each device separately which would need to be integrated into the GUI because that's where the device groups live. I'm not sure if there is a better, more general solution but it looks since there have been two papers on it, it's not as simple as it looks. |
|
Ok looks pretty good, ready for review https://35184-1301584-gh.circle-artifacts.com/0/dev/auto_tutorials/clinical/10_ieeg_localize.html |
 agramfort
left a comment
agramfort
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
just a nitpick
please merge if CIs are green
 larsoner
left a comment
larsoner
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Feel free to add Passing a schema to Validator around https://github.com/mne-tools/mne-python/blob/main/mne/conftest.py#L127 to get the CIs happy if you want!
add PR number fix flake put back test fix doc
sort of working version pretty much worked, could be cleaned up a bit though fix projection, add test more informative error, fix doc
|
Thanks @alexrockhill ! |
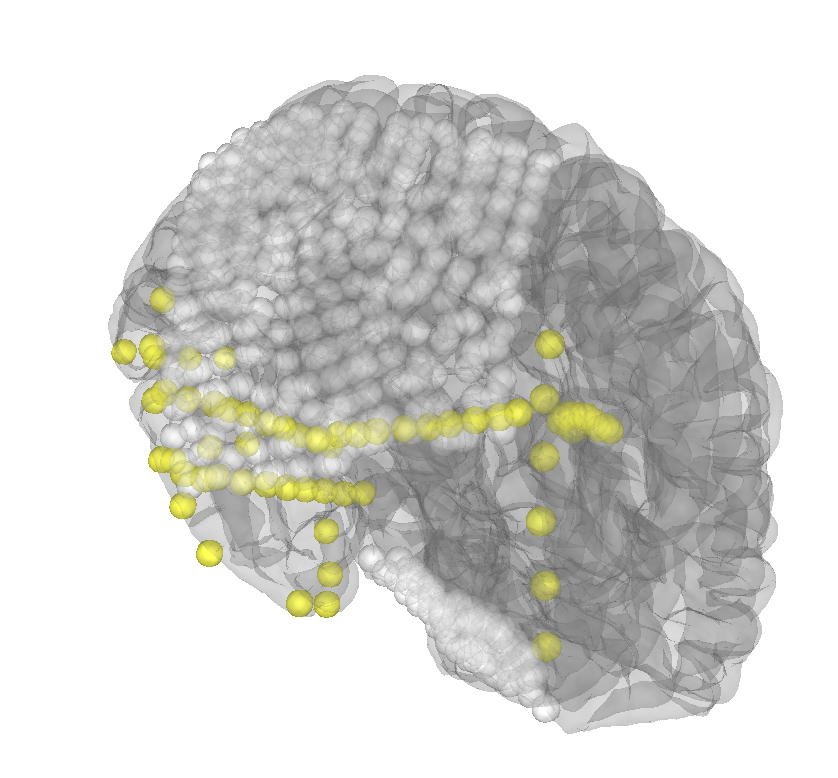
Add sensor projection to the brain surface to account for brain shift.