-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
MRG: Add Brain class based on _Renderer #6460
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
Most of the code comes from #6232 stripped from |
Codecov Report
@@ Coverage Diff @@
## master #6460 +/- ##
==========================================
- Coverage 89.27% 8.44% -80.83%
==========================================
Files 411 402 -9
Lines 74697 73403 -1294
Branches 12342 12248 -94
==========================================
- Hits 66684 6200 -60484
- Misses 5149 67148 +61999
+ Partials 2864 55 -2809 |
Codecov Report
@@ Coverage Diff @@
## master #6460 +/- ##
==========================================
- Coverage 89.27% 89.22% -0.05%
==========================================
Files 411 416 +5
Lines 74697 74371 -326
Branches 12342 12287 -55
==========================================
- Hits 66684 66356 -328
- Misses 5149 5154 +5
+ Partials 2864 2861 -3 |
|
Thanks to the discussion with the PyVista team, the latest fix allows us now to use the |
|
nice !
it works with add_data etc?
… |
|
I can't upload a GIF right now but basically the three examples I shared in this thread works so basic usage of |
|
How close are we then to having Maybe the plan should be:
WDYT? |
Not everything is available yet but we can definitely add the missing features incrementally. I can also create a RST table to improve visibility on this work. Benchmarking this
Sounds good to me! |
We probably do not need all of those examples, but we can go case by case. Let's aim to make sure
+1 for both. |
This PR adds the class |
|
Yes indeed! |
|
I can find a suitable value for the |
|
You can just add it to a todo list in a follow-up PR. It will quickly become annoying to test |
|
One thing that I'd like to do for now, though, is make it The reason for this is that we might want to:
Okay with you to keep it private for a bit @GuillaumeFavelier ? |
|
Yes sure, I'll rename it |
|
Thanks @GuillaumeFavelier, keep up the progress! |
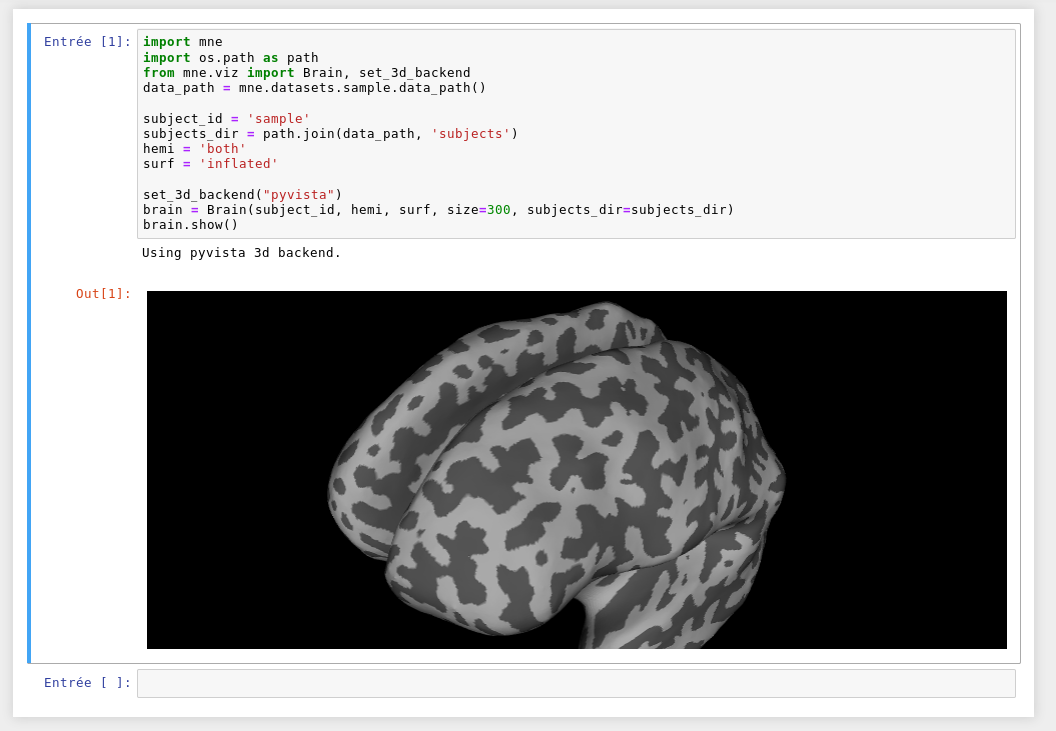

EDIT: In the last commit,
Brainhas been renamed into_Brainfrommne.viz._brain. The code snippets below are not up to date.While work is still in progress in #6232 to fix transparency issues, I thought it would be great to move forward the refactoring of the source code based on
PySurfer. This PR introduces the classBrainusing the 3d abstraction layer_Renderer.This PR is just the first step, the goal after is to refactor/test and replace parts of the source code relying on
PySurfer.Now I show some visualizations and code snippets using the latest 3d backend merged recently
PyVista:Basic Visualization
Activation Data Visualization
Display ROI Values