-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
[MRG, DOC] Added linear algebra of transform to doc #7087
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
Codecov Report
@@ Coverage Diff @@
## master #7087 +/- ##
==========================================
+ Coverage 89.74% 89.75% +<.01%
==========================================
Files 442 442
Lines 77786 77821 +35
Branches 12621 12633 +12
==========================================
+ Hits 69812 69845 +33
- Misses 5163 5166 +3
+ Partials 2811 2810 -1 |
| ax2.set_title('MRI Voxels') | ||
| ax3 = fig.add_axes([0.9, 0.1, 0.03, 0.8]) | ||
| fig.colorbar(plot, cax=ax3) | ||
| fig.show() |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
what is it you want to show here?
for making diagrams on sphinx I would look at https://sphinxcontrib-mermaid-demo.readthedocs.io/en/latest/
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Ohhhh I didn't put it together what Mainak wanted, I can do that
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
actually I've had trouble with sphinxcontrib-mermaid, it's not vigorously maintained (took several months for my PR to get merged, when all I was doing was bumping it to use the current stable version of mermaid). For the ICA tutorial I used graphviz (see here)
|
also the rendering is not working, something must be wrong with your rST: BTW if you do stick with mermaid, you need to add sphinxcontrib-mermaid to circle config and maybe also to doc/conf.py. If you search through the git history for "mermaid", you'll see that a few months ago there's a diff where I remove the relevant config lines. |
|
We now have some documentation here: @alexrockhill do you think it's useful to rework the PR here to fit into that or a complementary tutorial? FYI we release 0.21 in a couple of weeks, maybe this could make it in if we iterate a bit |
I'm just not sure how helpful this is... I like the idea but there's not a lot to visualize without mayavi or freesurfer plots. I don't even think the 2D images read in by nibabel like in the example you sent would really be all that helpful because it's 3D visualization. If I made images offline could they be added or is that too much bloat? |
We actually have 3D plotting abstraction in So maybe you can rework this PR using the abstracted functions? |
|
Ok so not that hard! Thanks for giving me a very nice solution basically ready to copy and paste in @larsoner! The one issue at this point is that the plotting of |
|
@larsoner, I did the coordinate transforms just like we did together for |
|
Thanks for working on this, seems like it'll be informative for people |
|
Ok I think this is ready. If you want to take a look @drammock or @agramfort, that would be great. |
|
CIs complain @alexrockhill legit? can you also share new rendered artifact when you have it? |
|
The CIs didn't look legit but four was a suspiciously large number to be failing, they were timing out though. Here's the latest tutorial page rendered: https://21868-1301584-gh.circle-artifacts.com/0/dev/auto_tutorials/source-modeling/plot_source_alignment.html#get-landmarks-in-head-space-from-the-digmontage-stored-in-raw |
|
I can take a look on Monday.
-------- Original Message --------
…On Aug 26, 2020, 13:59, Alex Rockhill wrote:
Ok I think this is ready. If you want to take a look ***@***.***(https://github.com/drammock) or ***@***.***(https://github.com/agramfort), that would be great.
—
You are receiving this because you were mentioned.
Reply to this email directly, [view it on GitHub](#7087 (comment)), or [unsubscribe](https://github.com/notifications/unsubscribe-auth/AAN2AU7ZL23UJDJLBM5NVWTSCVLRXANCNFSM4JPIZGQA).
|
 drammock
left a comment
drammock
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think this needs some work before it can be merged. The new material doesn't flow well with the old material around it and is a bit too terse/fragmentary (tutorials should tend toward being verbose and hand-holdy). I've left a number of more specific comments about that, as well as some stylistic comments about the code. I'll gladly review another iteration; don't hesitate to ping me for clarification / help.
| from scipy import linalg | ||
|
|
||
| import mne | ||
| from mne.datasets import sample |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
(nitpick) this always strikes me as unnecessary extra typing, since sample.data_path() is typically only used one time per tutorial.
| t1w = nib.load(op.join(data_path, 'subjects', 'sample', 'mri', 'T1.mgz')) | ||
| t1w = nib.Nifti1Image(t1w.dataobj, t1w.affine) | ||
| # XYZT_UNITS = NIFT_UNITS_MM (10 in binary or 2 in decimal) | ||
| # seems to be the default for Nifti files | ||
| # https://nifti.nimh.nih.gov/nifti-1/documentation/nifti1fields/nifti1fields_pages/xyzt_units.html | ||
| if t1w.header['xyzt_units'] == 0: | ||
| t1w.header['xyzt_units'] = np.array(10, dtype='uint8') | ||
| t1_mgh = nib.MGHImage(t1w.dataobj, t1w.affine) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'd like it if these lines were separated from the MNE-Python read_* lines, and were preceded by a prose explanation of what they're doing (or at least a single line comment to the tune of "now we'll use nibabel to load such-and-such MRI images and do whatever to them.")
I also don't understand the existing comment lines... you're changing the T1 header from 0 to 2, but you don't explain why it needs to be changed (in fact, because of the if-clause I don't even know for sure whether you're changing it just by reading/running the code). The best I can reconstruct is that 2 is the default, but I don't know why this file might be non-default, and what the consequences would be if I left it that way.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yeah, I didn't write the header stuff for this so I'm not sure.. The if is executed though so I'll fix that.
| # It is quite clear that the MRI surfaces (head, brain) are not well aligned | ||
| # to the head digitization points (dots). | ||
| # | ||
| # Compute the transform |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The placement of this new material seems odd to me. The preceding part of the tutorial used trans=None (so, no transform at all), and the very next line says "let's step through what the transform is doing". Maybe it would make more sense to put the new content after the "good example" section?
I also think the titles of the new sections are confusing. Here is what I think is going on; if this is right, maybe you can distill those bullets into better section titles (and also reduce heading level for the last 3, as implied here):
- visualizing the transformation of digitized points
- plot untransformed digitized points, as if they were in MRI RAS space
- apply the precomputed `trans` and plot again
- apply a second transform to get to "voxel space", and plot again
Note that "voxel" is not used anywhere else in the tutorial, so I think there is some work to be done integrating what you're trying to show here with the rest of the material (see also comment about variable names).
|
@drammock thanks so much for the comments, I was a bit too focused on the code working and should have given it a much better pass for writing, my bad! I made all the changes you suggested, and I think it's a lot better. |
|
@alexrockhill is this ready for a second review? |
|
@drammock yes please, I was waiting to see if the tests passed before I tagged you but looks like all good, ready to go, thanks!! |
|
Looks nice, I like all the changes 👍 |
|
@drammock feel free to merge once you and CircleCI are happy |
| vox_space = mne.transforms.apply_trans(ras2vox_tkr, meg_space * 1e3) # m → mm | ||
| mri_space = ((vox_space - 128) * [1, -1, 1])[:, [0, 2, 1]] * 1e-3 |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@alexrockhill I think the main source of confusion for me was that in your function you were referring to two coordinate systems as "ras" and "mri", which in MNE-Python (internally) those are called "mri" and "mri_voxel", respectively. That clears up the main confusion on my end, but there's still something I don't understand here: why are you applying a ras2vox transform to something that is in meg coordinate frame? Aren't we missing a step of transforming MEG frame to MRI Surface RAS first?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
If I'm not mistaken, the trans defines the transform from head to MRI surface RAS, combining the transformation from head to meg and meg to mri_ras so I think this should be changed to reflect that, but maybe let's check with @larsoner before going with that answer
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
In that case we shouldn't call that variable meg_space, and some of the prose needs to change.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yeah that's how it was before and then I changed it to meg_space to match from mri_space but I don't think that was right
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Ok I changed it the way I think it should be
|
@alexrockhill I confess I'm still confused. As far as I can tell, the |
|
all FreeSurfer surfaces (including seghead) are in the "MRI" (FreeSurfer surface RAS) coordinate frame in |
|
|
|
after an offline chat with @larsoner about the big-picture goals here, I made some radical edits to the new material to (hopefully) distill this section down to what (I think?) are the main insights we're trying to teach people here: how the coord systems differ, and how to convert points from one to another coord sys. @alexrockhill please take a look and let me know if you think this leaves out anything from the old version that you think is important for users to know. |
|
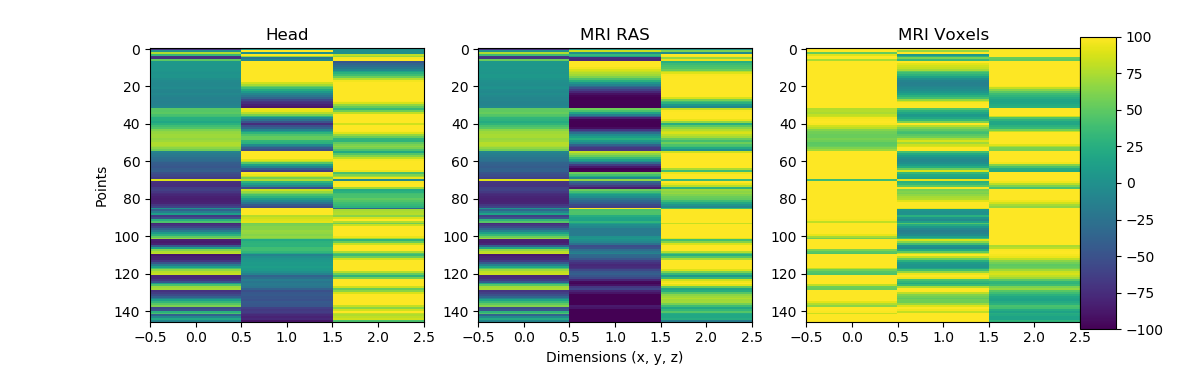
It definitely distills the information and I like that, and you can understand the mri voxel coordinate frame as a person lying supine as the slices are acquired in the z-plane. The code is great, and I think super helpful. The two areas for improvement I see if that the one figure is very busy although efficient in conveying information, I think the transparency could be turned down or it could be broken into one or two other figures. Second is crucial step (1/2) for anyone who might use this is applying the trans to go from head to mri (step (2/2) is the voxel transformation), and I think that could be added, even in commented out code would be fine I think. |
|
I went to go make those changes and I realized I was just putting everything back how it originally was so I'm going to stop. I'm fine with how it is, although I'm not sure it accomplishes the original goal I had of explaining how to transform points that an end-user would have in head space for any number of reasons and want to transform them to mri or mri_voxel space. They could definitely use the wrapped functions and never need to know anything about these transforms but for me, they were very useful to know about for translating points to do defacing, changing coordinate frames for stereo eeg and probably also a project plotting the location of a TMS coil coming up. I don't think this tutorial addition conveys the information that I would have needed to do the deface algorithm although it does add novel information about the voxel space. I like the additional information but I feel like the core information that I was trying to pass forward to the next person in my position isn't there or is only there if you take the inverse of all this tutorial addition and then somehow know to skip the extra step of going through meg space. I started this PR because Eric asked me to/suggested it and to pass forward the information that he explained to me and I think the new version accomplishes a different goal. I know the information now so it doesn't really effect me so I thought I would give a verbose explanation on my opinion but it's really something I'd defer on about what this tutorial is to contain. |
|
@alexrockhill you've convinced me that it's worthwhile to show both directions of transformation. I've added back an example of head -> mri -> voxel space, simplified to use just the nasion point rather than the full set of digitized points. |
|
Looks great! I don't know how easy it is to reference |
You can push a commit to do that now, or you can do it in a separate PR. It looks like mne-bids does publish an objects.inv file, so it shouldn't be too painful: add mne-bids URL to the relevant part of |
|
I might do that in a separate PR... Thanks for pointing the right direction! |
|
OK: I'm happy, Alex is happy, and Circle is happy. Merging! |
|
Very cool thanks for all the help!! And thanks Eric too! |
|
great team effort https://www.youtube.com/watch?v=g1EEVNXaKvA ! :) |
* upstream/master: (489 commits) MRG, DOC: Fix ICA docstring, add whitening (mne-tools#8227) MRG: Extract measurement date and age for NIRX files (mne-tools#7891) Nihon Kohden EEG file reader WIP (mne-tools#6017) BUG: Fix scaling for src_mri_t in coreg (mne-tools#8223) MRG: Set pyvista as default 3d backend (mne-tools#8220) MRG: Recreate our helmet graphic (mne-tools#8116) [MRG] Adding get_montage for montage to BaseRaw objects (mne-tools#7667) ENH: Allow setting tqdm backend (mne-tools#8177) [MRG, IO] Persyst reader into Raw object (mne-tools#8176) MRG, BUG: Fix errors in IO/loading/projectors (mne-tools#8210) MAINT: vectorize _read_annotations_edf (mne-tools#8214) FIX : events_from_annotation when annotations.orig_time is None and f… (mne-tools#8209) FIX: do not project to sphere; DOC - explain how to get EEGLAB-like topoplots (mne-tools#7455) [MRG, DOC] Added linear algebra of transform to doc (mne-tools#7087) FIX: Travis failure on python3.8.1 (mne-tools#8207) BF: String formatting in exception message (mne-tools#8206) BUG: Fix STC limit bug (mne-tools#8202) MRG, DOC: fix ica tutorial (mne-tools#8175) CSP component order selection (mne-tools#8151) MRG, ENH: Add on_missing to plot_events (mne-tools#8198) ...
* added linear algebra of transform to doc trying to fix circle bad plot oops forgot titles overlaps trying mermaid * working 3D plotting, RAS conversion is wrong * another try at coordinate frames * fix html rendering * nice suggestion Eric * changes didn't save * fix broken ref * added better comments * more comments * alex review comments * added a bit to latest * Dan comments * mayavi fig -> older version figure * fix ref error * a few more small word changes to align verbage * unnecessary copy * simplify function * Update tutorials/source-modeling/plot_source_alignment.py * Dan's suggestions * put back renderer * added scene * fix renderer * nevermind that's wrong * smooth prose, simplify function * fix circle * meg -> mri * more prose improvements; explain unit conversion * fix codespell * a fresh take * restore example going from head to vox * fix focalpoint * fix view again Co-authored-by: Alex <aprockhill206@gmail.com> Co-authored-by: Daniel McCloy <dan@mccloy.info> Co-authored-by: Eric Larson <larson.eric.d@gmail.com>
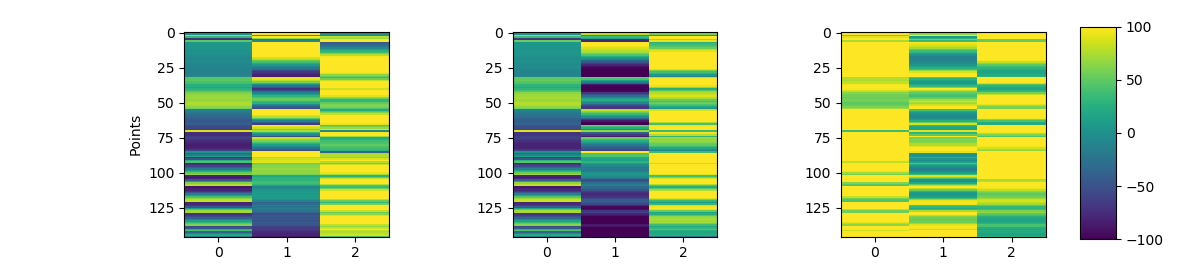

Ok here is the linear algebra of how the transform happens. I think it's useful for developers but it's a bit hard to visualize without putting up some mayavi/matplotlib 3D figures with just points on them which would be far below the rest of the example quality-wise. If there is a way to plot it that looks nice, I could add that. I'm not sure if it'd be useful to print details about the scaling or anything but it does explain how I understand the transform to work. If that's helpful to have in docs then great, if this isn't good enough to be merged that would be understandable too.
Addresses #7081