-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
FIX: do not project to sphere; DOC - explain how to get EEGLAB-like topoplots #7455
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
This already looks reasonable to me. I'm not too worried about the realistic positions looking bad (as above at least) on a sphere as long as they do so consistently, they probably shouldn't look perfect. |
|
If once you're done @mmagnuski can you push a commit with |
|
My priority is that theoretical locations look good on the 2D topomap projection. I don't think it is necessary to make realistic locations look good on a sphere, or the other way round (theoretical locations on a realistic head). |
|
@larsoner @cbrnr |
|
just letting you know that I'll be back to it tomorrow, I'll have to figure out what should be happening now in the lasso selection test (some of the steps in the test are not super-clear). |
|
@mmagnuski any chance to complete this quickly before the imminent 0.20 release? |
|
@agramfort Depends how imminent is that release :) |
|
reallly imminent like by the end of the week for sure
… |
|
how can we help?
… |
That would be great, when you're happy please push a commit with
It only took about 20 minutes to look through last time I think. I would say if nothing is obviously broken we merge, if it breaks a lot of stuff we push to 0.21 (or maybe 0.20.1 via backport). |
|
@agramfort I just have to fix tests, I had a problem with lasso selection test - but I have solved it now, thanks for reminding me about this issue! @larsoner Ok, that sounds good. :) I'm not sure about raw = read_raw_fif(raw_fname, preload=True)
sizes = {'mag': 828, 'grad': 1700, 'eeg': 386}
nchans = {'mag': 102, 'grad': 204, 'eeg': 60}
ch_type = 'eeg'
conn, ch_names = find_ch_connectivity(raw.info, ch_type)
# Silly test for checking the number of neighbors.
assert_equal(conn.getnnz(), sizes[ch_type])the expected number of non-zero elements for eeg is |
Codecov Report
@@ Coverage Diff @@
## master #7455 +/- ##
==========================================
- Coverage 90.07% 89.73% -0.34%
==========================================
Files 454 454
Lines 82353 81536 -817
Branches 13016 13015 -1
==========================================
- Hits 74180 73170 -1010
- Misses 5350 5503 +153
- Partials 2823 2863 +40 |
|
Yes I think that's reasonable |
|
Ok, one test is still broken, I'll fix it in the evening and then we'll see |
|
My vote is to push this to 0.21, then. I don't see this as a critical release blocker and we don't have consensus on a quick solution |
|
I agree move to 0.21 unless there is something we agree on in the next 24h
before the release
… |
|
I agree, I was actually expecting this, that's why I thought this might take longer.
These are partly overlapping issues, but the main thing this PR fixes is not the problems you previously mentioned about the layout of standard montages but #7341. The problem was that using
This is what I previously meant about choosing the default sphere radius so that channel layouts look good by default. This definitely requires some thought and time. |
|
@cbrnr |
|
@larsoner @agramfort |
|
Ok
|
| # In mne-python the head center and therefore the sphere center are calculated | ||
| # using fiducial points. Because of this the head circle represents head |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This seems a bit misleading. The center of the head coordinate frame is defined by the fiducial points. The automated sphere center is calculated using all EEG + fiducials + extra digitization points. With this wording, people might think both of these are calculated just from the three fiducial points. Maybe something like:
| # In mne-python the head center and therefore the sphere center are calculated | |
| # using fiducial points. Because of this the head circle represents head | |
| # In mne-python the head coordinate frame is defined by the fiducial points. | |
| # (LPA, Nasion, RPA). Because of this the head circle represents head |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, that's better, thanks. But it seems I may still not understand something here - I'm not sure what you are referring to as "the automated sphere center". The default sphere center is (0, 0, 0), right?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The docs say:
The sphere parameters to use for the cartoon head. Can be array-like of shape (4,) to give the X/Y/Z origin and radius in meters, or a single float to give the radius (origin assumed 0, 0, 0). Can also be a spherical ConductorModel, which will use the origin and radius. Can be “auto” to use a digitization-based fit. Can also be None (default) to use ‘auto’ when enough extra digitization points are available, and 0.095 otherwise. Currently the head radius does not affect plotting.
So for mon.plot in theory there should be enough points (EEG points can be used) and so the origin should not be (0, 0, 0). If it is, we either have an implementation (probably this) or a documentation bug.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
at least I think it will try to use the EEG dig points, even though it says "extra digitization points"...
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Okay I was wrong, it does just look for extra:
Line 650 in af1120c
| get_fitting_dig(info, 'extra', verbose='error') |
IIRC the motivation was that when you apply a montage, missing channels are not included from info['dig']. I think if we kept them all in there (maybe with the others as extra points?) then checked for EEG+extra, we could make it always fit an origin.
We can look into enabling this -- I actually don't think it would be too much work in DigMontage -- but if what you have here works well enough already, then we should just stick with it (especially right before release) rather than try to make DigMontage do more.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Defining head center (and sphere origin) based on fiducials is simple and makes sense. However, I'm not so sure about using all available dig and channel points for that: because then, depending on the distribution of these points, the head outline has a different meaning. I think it is good to have a consistent meaning for what the head outline represents. But I may still be missing something here.
| # calculate the sphere origin from position of Oz, Fpz, T3/T7 or T4/T8 | ||
| # channels. This is easier once the montage has been applied to the data and | ||
| # channel positions are in head space - see this example | ||
| # (all the code below should be a separate example): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Feel free to make a separate examples/visualization/plot_eeglab_head_sphere.py or so, then link to it here
|
|
||
| n_channels = len(biosemi_montage.ch_names) | ||
| fake_info = mne.create_info(ch_names=biosemi_montage.ch_names, sfreq=250., | ||
| ch_types=['eeg'] * n_channels) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
ch_types='eeg' works and is shorter
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Ah, great!
| fake_raw.set_montage(biosemi_montage) | ||
|
|
||
| check_ch = ['Oz', 'Fpz', 'T7', 'T8'] | ||
| pos = fake_raw._get_channel_positions(picks=check_ch) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
shouldn't use a private function in an example so we have to wait for #7667
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Oh, yes, the example part is sketchy for now. Great to know .get_ch_positions() will be added, I was using my own function get_ch_pos() for years! :)
Co-authored-by: Eric Larson <larson.eric.d@gmail.com>
Co-authored-by: Eric Larson <larson.eric.d@gmail.com>
| # positions onto a sphere. Because ``'standard_1020'`` montage contains | ||
| # realistic, not spherical, channel positions, we will use a different montage | ||
| # to demonstrate controlling how channels are projected to 2d space. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
just a fly-by comment --> many users (including myself) will want to "fix" the plotting of 'standard_1020' and not the biosemi montage. Can that be included in this brief example? :)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This PR seems 98% ready to go (right @mmagnuski ?), in which case let's save that for #7141 unless it's trivial to add here
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Unfortunatelly the standard_1020 is not spherical, it contains realistic channel positions, I don't think it can be easily fixed. The easiest fix would be to distribute the spherical 1020 locations, which is discussed here: #7141 (as you know, for sure, cause you took part in the discussions :) ). Once this is done I'd happily change the tutorial to use the spherical 1020.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
alright, thanks - I guess I was hoping against all odds to find a ready-made solution here. Thanks for your PR!
|
|
||
| fig.texts[0].remove() | ||
| ax[0].set_title('MNE channel projection', fontweight='bold') | ||
| ax[1].set_title('EEGLAB channel projection', fontweight='bold') |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Also looks like these did not render, probably because the axes are turned off
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Try in native Python instead? I would try tight_layout, subplots_adjust, or if you're brave, constrained_layout
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The images on circle look cropped at the top compared with what I get locally in the notebook. Maybe the titles go outside figure - IIRC notebook usually extends the rendered space in such cases, but the saved image is still cropped (meaning: not extended). I'll add top gridspec_kws in the example then.
Co-authored-by: Eric Larson <larson.eric.d@gmail.com>
Co-authored-by: Eric Larson <larson.eric.d@gmail.com>
| # ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ | ||
| # | ||
| # Channel positions in 2d space are obtained by projecting their actual 3d | ||
| # positions using a sphere as a reference. Because ``'standard_1020'`` montage |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@larsoner I changed this to "using a sphere as a reference" - this may be less clear, but is closer to what we do. After all it was funny to mention that channels are projected to a sphere in a PR titled "do not project to sphere" 😀
|
The links to the tutorial and the example: The remaining TODOs (feel free to add your comments to be adressed here):
|
|
@larsoner @agramfort
This might be confusing to some people as topoplot outside of mne is commonly associated with what is known as topomap in mne. However usage of layouts has been deprecated in topomaps. |
 drammock
left a comment
drammock
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
this looks great. I've made a bunch of small nitpicky suggestions, but generally I think it's very clear and helpful.
| # make x and limits the same in both panels | ||
| ylm = ax[1].get_ylim() | ||
| xlm = ax[1].get_xlim() | ||
| ax[0].set_ylim(ylm) | ||
| ax[0].set_xlim(xlm) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
passing sharex and sharey to plt.subplots above should make this unnecessary
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Oh, right, thanks!
| # make x and limits the same in both panels | ||
| ylm = ax[1].get_ylim() | ||
| xlm = ax[1].get_xlim() | ||
| ax[0].set_ylim(ylm) | ||
| ax[0].set_xlim(xlm) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
sharex / sharey again
Co-authored-by: Daniel McCloy <dan@mccloy.info>
|
Thanks for all the suggestions @drammock ! |
 larsoner
left a comment
larsoner
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
LGTM @drammock feel free to merge if you're happy
|
thanks @mmagnuski! |
* upstream/master: (489 commits) MRG, DOC: Fix ICA docstring, add whitening (mne-tools#8227) MRG: Extract measurement date and age for NIRX files (mne-tools#7891) Nihon Kohden EEG file reader WIP (mne-tools#6017) BUG: Fix scaling for src_mri_t in coreg (mne-tools#8223) MRG: Set pyvista as default 3d backend (mne-tools#8220) MRG: Recreate our helmet graphic (mne-tools#8116) [MRG] Adding get_montage for montage to BaseRaw objects (mne-tools#7667) ENH: Allow setting tqdm backend (mne-tools#8177) [MRG, IO] Persyst reader into Raw object (mne-tools#8176) MRG, BUG: Fix errors in IO/loading/projectors (mne-tools#8210) MAINT: vectorize _read_annotations_edf (mne-tools#8214) FIX : events_from_annotation when annotations.orig_time is None and f… (mne-tools#8209) FIX: do not project to sphere; DOC - explain how to get EEGLAB-like topoplots (mne-tools#7455) [MRG, DOC] Added linear algebra of transform to doc (mne-tools#7087) FIX: Travis failure on python3.8.1 (mne-tools#8207) BF: String formatting in exception message (mne-tools#8206) BUG: Fix STC limit bug (mne-tools#8202) MRG, DOC: fix ica tutorial (mne-tools#8175) CSP component order selection (mne-tools#8151) MRG, ENH: Add on_missing to plot_events (mne-tools#8198) ...
…opoplots (mne-tools#7455) * FIX: do not project to sphere * FIX: fix tests * FIX: check if border is correct before _get_extra_points * DOC: add controlling channel topo projection to sensor locations tutorial * better option checking Co-authored-by: Eric Larson <larson.eric.d@gmail.com> * fix missing space Co-authored-by: Eric Larson <larson.eric.d@gmail.com> * use Eric's suggestion - format string Co-authored-by: Eric Larson <larson.eric.d@gmail.com> * use Eric's suggestion - random seed Co-authored-by: Eric Larson <larson.eric.d@gmail.com> * DOC: add example and link the tutorial and the example * FIX: fix syntax and spelling in the example * FIX: fix example * FIX: more fixes, the example and one topomap test * FIX: and some more fixes to the example * Apply suggestions from Daniel's code review Co-authored-by: Daniel McCloy <dan@mccloy.info> * DOC: use sharex, sharey, as suggested by @drammock, also fix tutorial thumbnail * FIX: fix links in the example Co-authored-by: Eric Larson <larson.eric.d@gmail.com> Co-authored-by: Daniel McCloy <dan@mccloy.info>
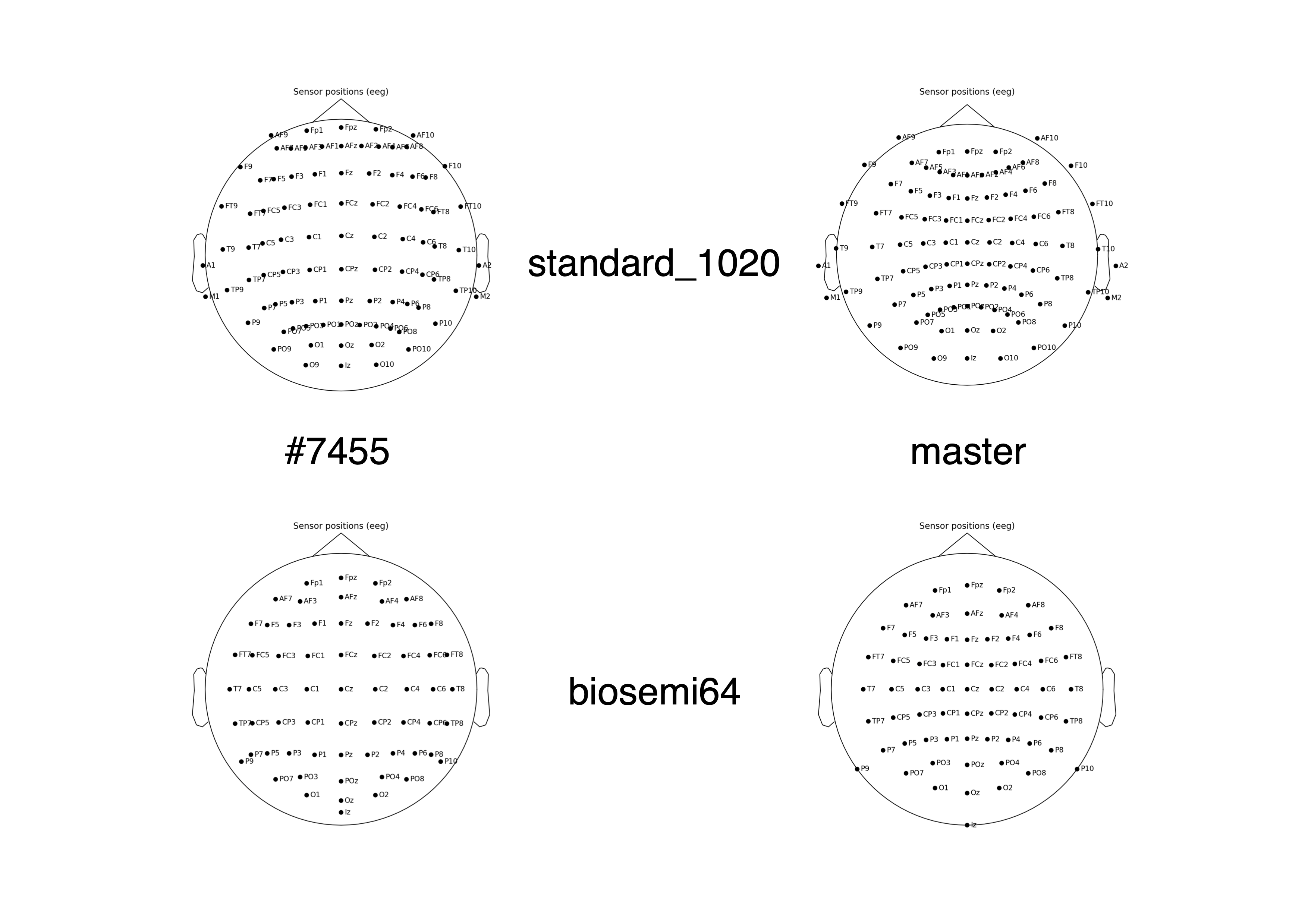
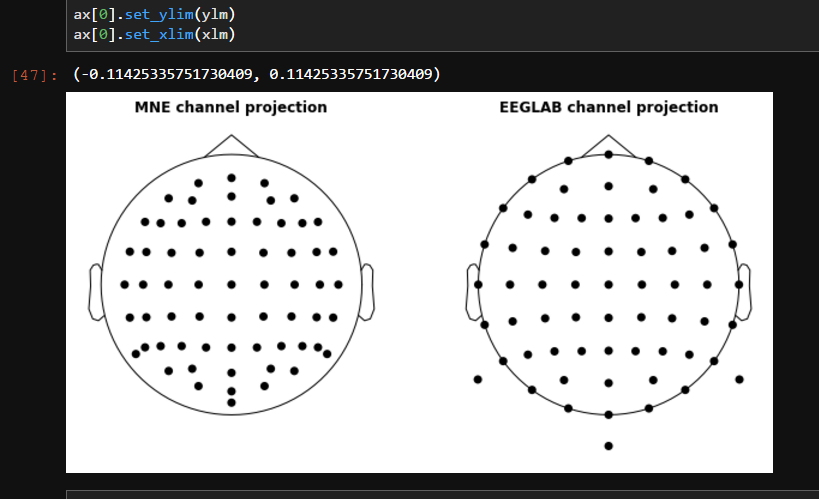
Fixes #7341
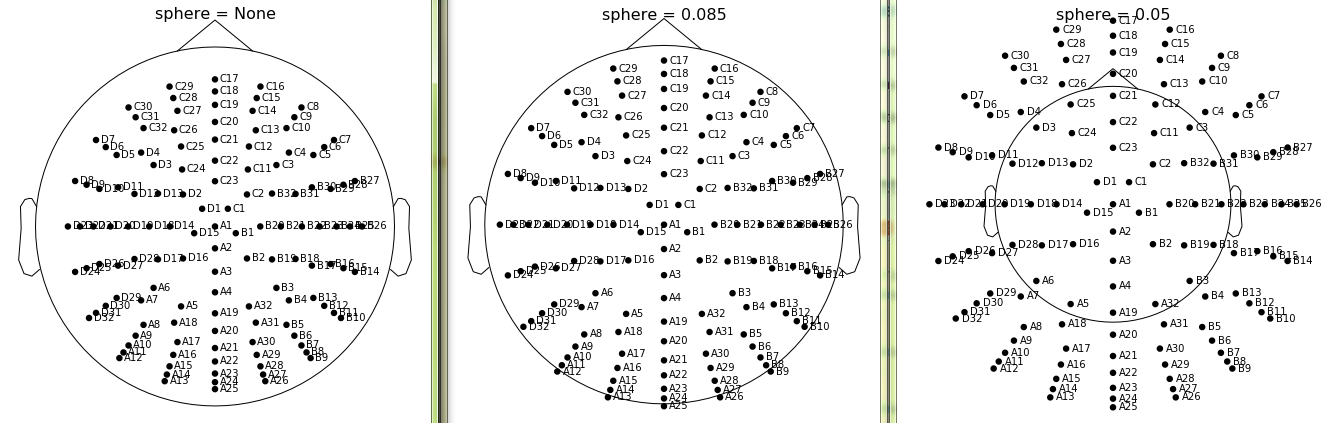
Currently all channels are projected to a sphere so the sphere radius does not take any effect. If we don't project to a sphere the sphere radius changes the channel positioning with respect to head outline:
However this means that channel positions that are not spherical, like EGI (or standard_1020) do not look so good because we no longer project to sphere.
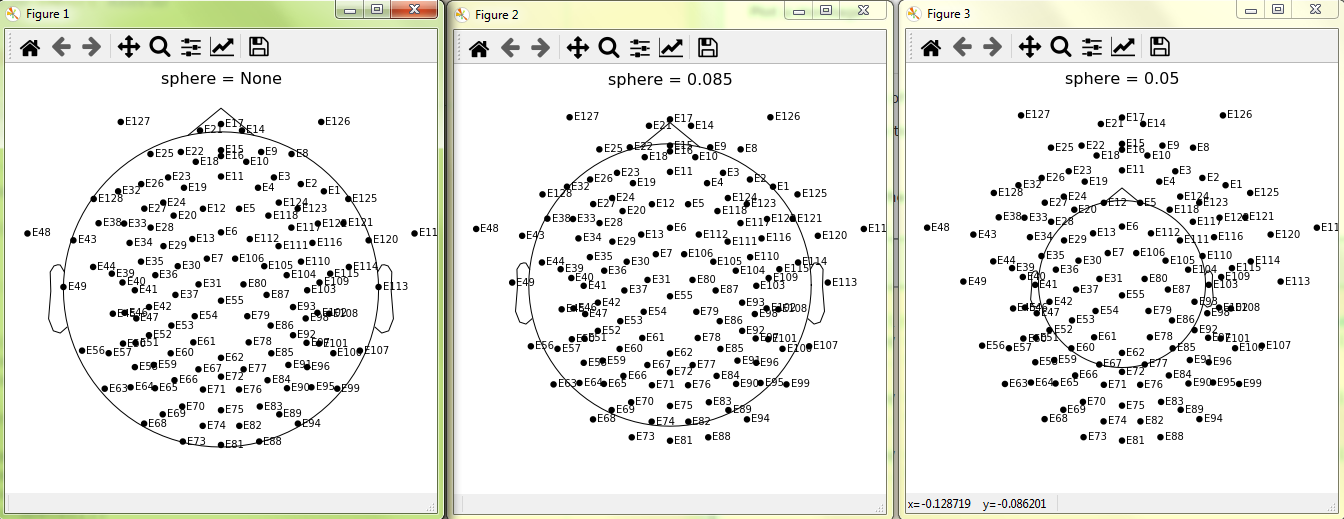
One way to circumvent this would be to project to sphere and then scale channel positions by the average channel pos radius (currently each channel is scaled by its own radius, so the channels are no longer on a sphere).