-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
MRG, FIX: Fix MRI orientations #7725
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
Codecov Report
@@ Coverage Diff @@
## master #7725 +/- ##
==========================================
- Coverage 90.24% 90.23% -0.01%
==========================================
Files 455 455
Lines 84626 84698 +72
Branches 13401 13421 +20
==========================================
+ Hits 76367 76429 +62
- Misses 5392 5402 +10
Partials 2867 2867 |
 agramfort
left a comment
agramfort
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
+1 for MRG if @christian-oreilly confirms it fixes his pb
|
friendly ping to @christian-oreilly |
|
thx @larsoner |
|
@larsoner Sorry for the late feedback. I do not have the MRI that prompted me to submit the issue #7221 anymore because I went around the issue by preprocessing all my MRI to a "conform" space in the meantime. But I always end up having to move data between pipelines and almost always run into issues with incompatible coordinate systems. So I found some MRI from a different project (originally processed in CIVET rather than FreeSurfer) that were not preprocessed to a conform to space and that is what I got: So... the fix works to the extent that it puts the MRI in coronal view when it is supposed to put it in this view. I think the rest is more about what level of user-friendliness you want to support (i.e., whether or not you would like to have it 100% aware of coordinate system so that it would invert axes if needed to display it in a standard orientation or leave it to the user to provide their MRI in a given coordinate system). At least, right now, it does the volume along the requested axis. FYI, there appears to be some bug in the management of paths: The call at "In [9]" should works as, according to the docstring, the subjects_dir argument can be left to None, in which case "the path is obtained by using the environment variable SUBJECTS_DIR." This error seems to have been introduced with this fix because it works fine when I revert to the MNE version I had before applying this fix. |
|
Indeed I can see how this error was created. I can fix the flipping problem more easily if you can share their T1.mgz and |
|
@larsoner Sure, I'll sent these to you. |
|
@christian-oreilly can you first try #7763 ? I think I only need the files if that PR does not work. |
|
@larsoner Sure, I'll do. |
|
@larsoner Yep, works fine! Both the orientation and the path thing. Nice work! :) |
* upstream/master: (74 commits) FIX: Correct a bug in find_bads_eog (mne-tools#7797) [MRG] split_naming='bids' changes from _part-%d to _split-%d (mne-tools#7794) MRG, MAINT, DOC: Remove spyder (mne-tools#7796) MAINT: fixes for linkcheck (mne-tools#7762) [WIP] Update ieeg data example for ECoG (mne-tools#7768) fix examples/tutorials [circle full] (mne-tools#7786) MAINT: Clean up VTK and add to pre on Azure (mne-tools#7780) ENH: Add matplotlib animation support [skip travis] (mne-tools#7783) MRG, API: change out_type default in permutation_cluster_(1samp_)test (mne-tools#7781) DOC: docstring fixes (mne-tools#7777) MRG, ENH: Add tol_kind option (mne-tools#7736) MRG, DOC: Notes about info (mne-tools#7772) ENH: Speed up NIRx read without preload (mne-tools#7759) Minor plot_raw aes improvement (mne-tools#7770) MRG, FIX: Fixes for BEM contours (mne-tools#7763) MRG, STY: Fix E741 (mne-tools#7767) MRG, ENH - Plot optodes in plot_alignment for fNIRS channels (mne-tools#7747) FIX: Update NIH support [skip travis] (mne-tools#7766) MAINT: Bump tol for gamma map test (mne-tools#7764) MRG, FIX: Fix MRI orientations (mne-tools#7725) ...
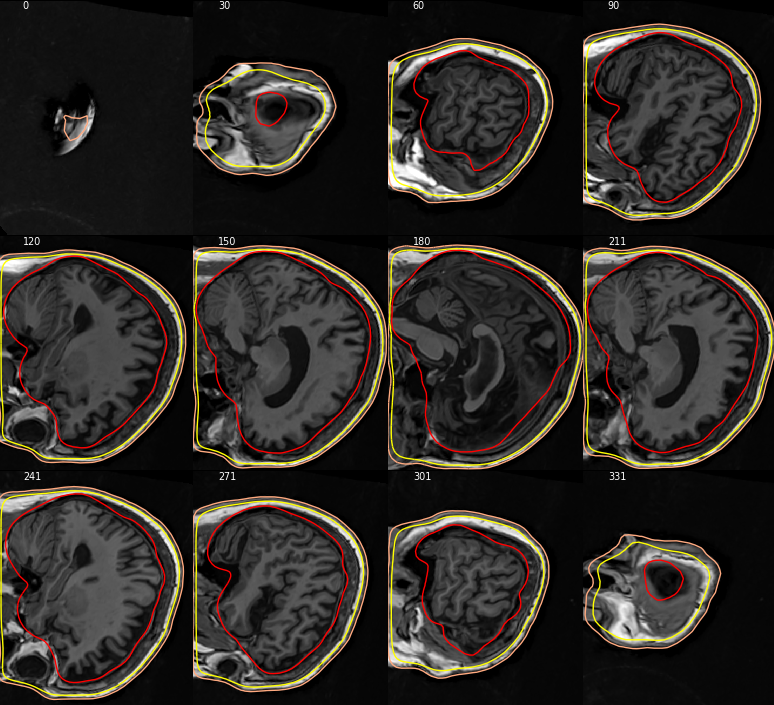
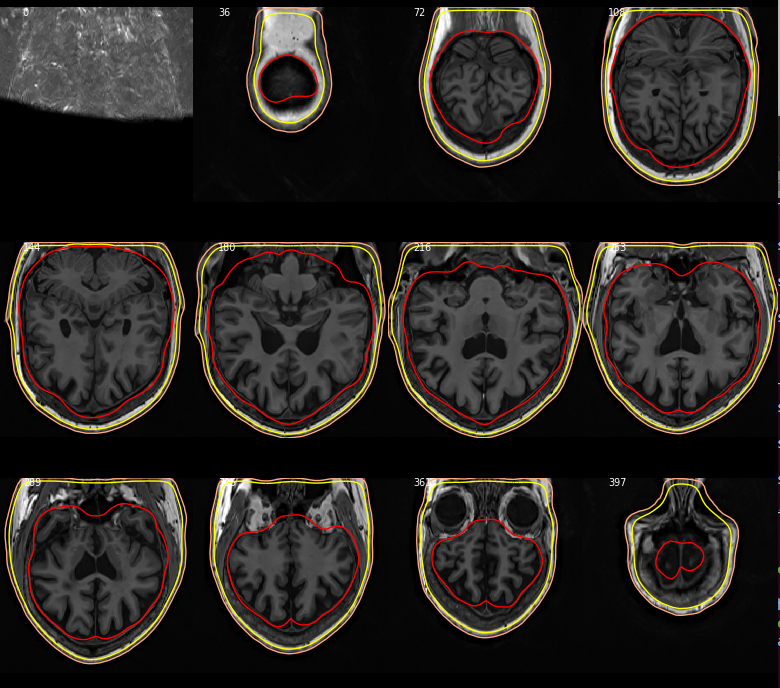
Closes #7221.
@christian-oreilly can you try it with your MRIs that are in non-standard orientation? I was able to check that the FreeSurfer-standard ones look okay, but don't have good MRIs to test for non-standard orientations. If you don't have them I could maybe resample these and regenerate BEMs but that sounds like too much work :)
This actually makes our code simpler and easier to read I think, which is nice.