-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
MRG, FIX: Fixes for BEM contours #7763
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Merged
Merged
Conversation
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
Member
Author
|
Reported as fixing orientations for @christian-oreilly here, so this one should be good to go |
Member
|
thanks for cleaning my mess @larsoner :) |
Member
Author
Member
Author
Member
Author
|
Style failure is just the one from #7767 |
agramfort
approved these changes
May 12, 2020
Member
|
thx @larsoner |
larsoner
added a commit
to larsoner/mne-python
that referenced
this pull request
May 19, 2020
* upstream/master: (74 commits) FIX: Correct a bug in find_bads_eog (mne-tools#7797) [MRG] split_naming='bids' changes from _part-%d to _split-%d (mne-tools#7794) MRG, MAINT, DOC: Remove spyder (mne-tools#7796) MAINT: fixes for linkcheck (mne-tools#7762) [WIP] Update ieeg data example for ECoG (mne-tools#7768) fix examples/tutorials [circle full] (mne-tools#7786) MAINT: Clean up VTK and add to pre on Azure (mne-tools#7780) ENH: Add matplotlib animation support [skip travis] (mne-tools#7783) MRG, API: change out_type default in permutation_cluster_(1samp_)test (mne-tools#7781) DOC: docstring fixes (mne-tools#7777) MRG, ENH: Add tol_kind option (mne-tools#7736) MRG, DOC: Notes about info (mne-tools#7772) ENH: Speed up NIRx read without preload (mne-tools#7759) Minor plot_raw aes improvement (mne-tools#7770) MRG, FIX: Fixes for BEM contours (mne-tools#7763) MRG, STY: Fix E741 (mne-tools#7767) MRG, ENH - Plot optodes in plot_alignment for fNIRS channels (mne-tools#7747) FIX: Update NIH support [skip travis] (mne-tools#7766) MAINT: Bump tol for gamma map test (mne-tools#7764) MRG, FIX: Fix MRI orientations (mne-tools#7725) ...
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Add this suggestion to a batch that can be applied as a single commit.
This suggestion is invalid because no changes were made to the code.
Suggestions cannot be applied while the pull request is closed.
Suggestions cannot be applied while viewing a subset of changes.
Only one suggestion per line can be applied in a batch.
Add this suggestion to a batch that can be applied as a single commit.
Applying suggestions on deleted lines is not supported.
You must change the existing code in this line in order to create a valid suggestion.
Outdated suggestions cannot be applied.
This suggestion has been applied or marked resolved.
Suggestions cannot be applied from pending reviews.
Suggestions cannot be applied on multi-line comments.
Suggestions cannot be applied while the pull request is queued to merge.
Suggestion cannot be applied right now. Please check back later.
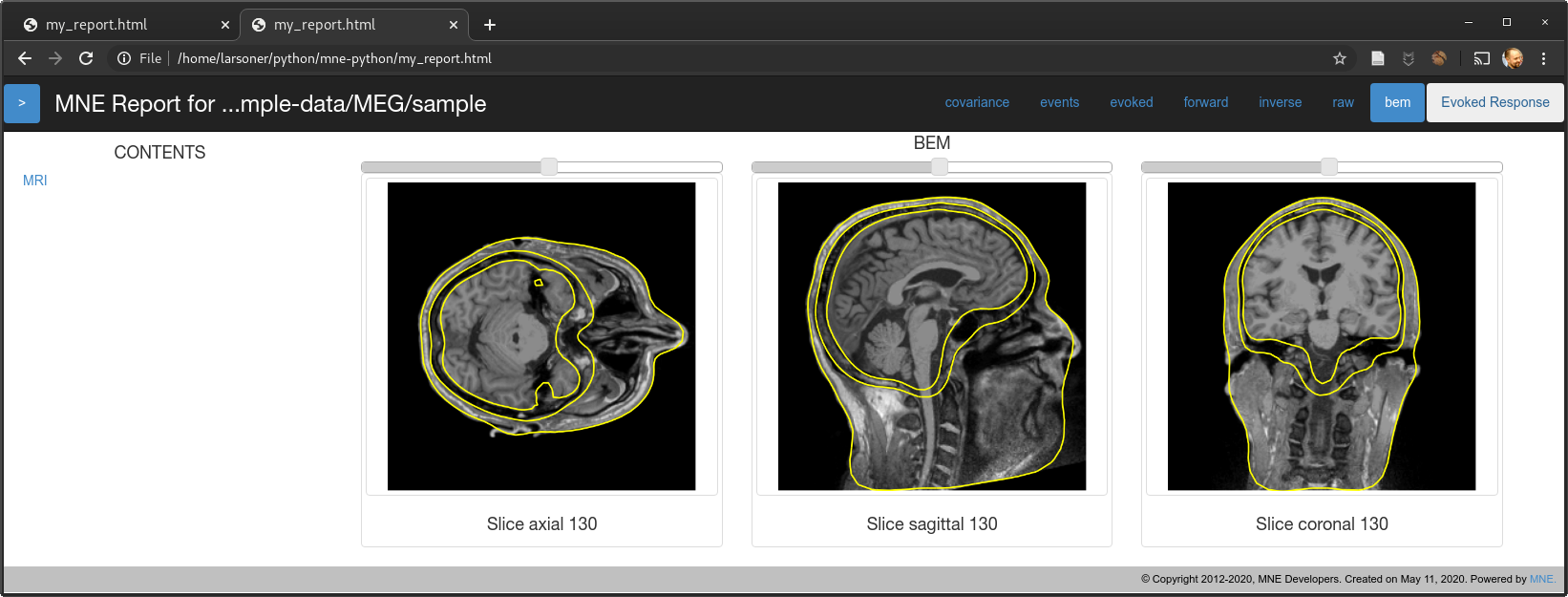
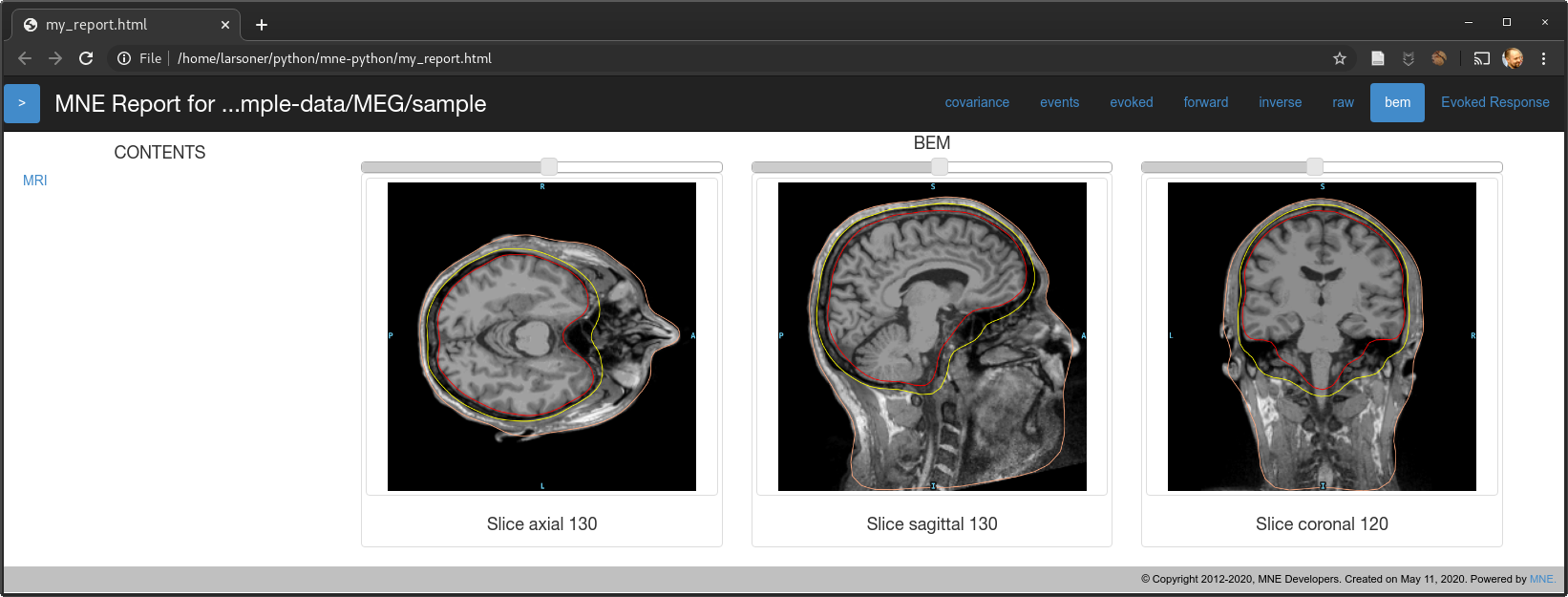
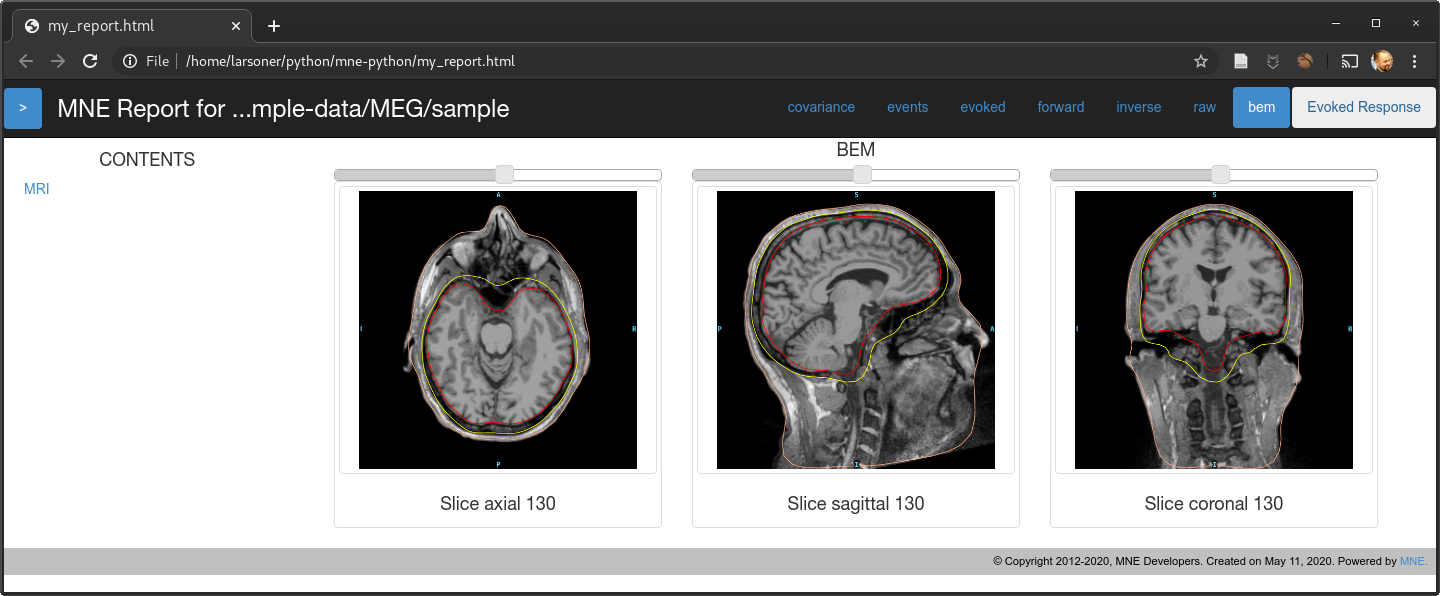
Should fix the bug mentioned in #7725. @christian-oreilly can you try it with your out-of-standard-ori MRI? I don't have any to test with but in principle I think this should work...
This PR also:
show_orientations=False)mne.viz._3d._plot_mri_contoursthat we should really refactor with themne.viz.misc._plot_mri_contours-- the function names match and there is a lot of similar code so it's unfortunate that we ended up with two copies, presumably due to @agramfort 6 years ago accidentally making two copies in 5c1d64b :)slices=Nonefrom:masterwe correctly avoid the last but not the first and unevenly sample in between, we now evenly sample from near-to-first to near-to-last instead by making 14 points evenly spaced between first and last slice (inclusive) and then excluding the first and last of those 14 to obtain 12.On master for
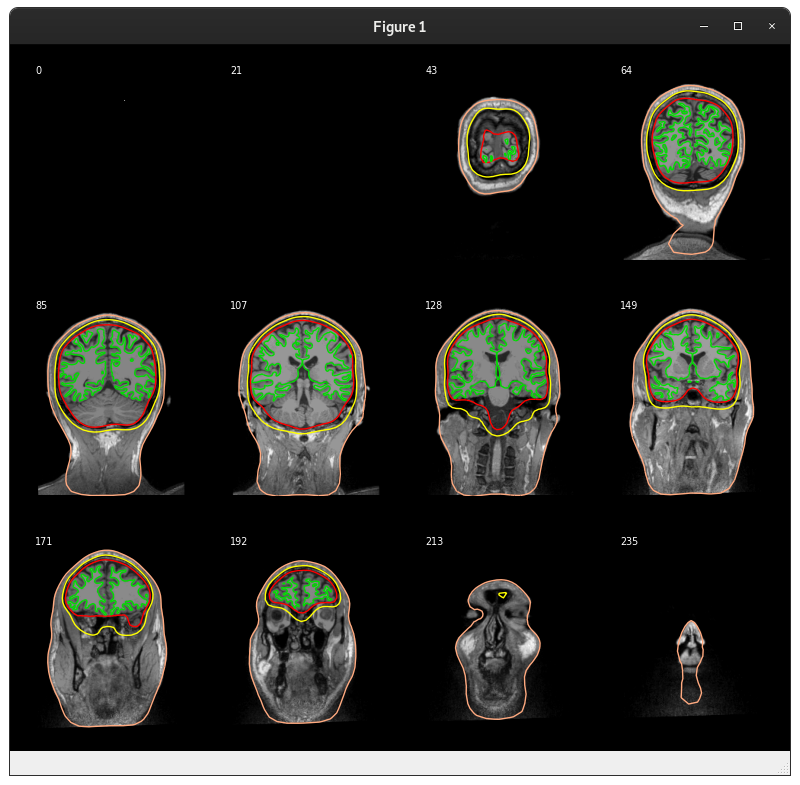
plot_forward.pywe get:And on this PR:
Note that there is a left-to-right flip here. On
masterthe subject's left was shown on the right (radiological convention, probably not intentional) and this PR sets it to the more standard (for us at least) neurological convention of having the subject's left on the left. You can tell this by the fact that in the image from this PR, slice 177 (bottom left) has the pill on the right, and then the last slice is flipped left-right relative to themasterplots.