-
-
Notifications
You must be signed in to change notification settings - Fork 1.5k
WIP: Compare Beer-lambert to HOMER #8711
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
Hi Eric, Wow I remember digging in to the beer Lambert law in great detail when this code was written. I thought at the time I had documented it well, but looking back I can barely remember what I did 😫. I will list my recollections below and where Im guessing differences might be sneaking in, but I am away from my computer at the moment so can't check these things myself, but once I am back at work next week I can chase this up with you. Each software uses different defaults for d/ppf, but I would have expected if we used the same value then the results would be similar. We should definitely understand what's going on here. A factor of 5 difference in ppf to get the same result is too much (and 5 is a fishy number, and not one that I recall being in the code, so possibly multiple things underly the difference you are observing?). First off, I validated this implementation against the NIRS-Toolbox and not Homer. But I remember eye balling the Homer code at the time. The script I used to validate our implementation is here https://github.com/rob-luke/mne-fnirs-demo/blob/master/BeerLambert_Validation.ipynb. I recall there were some differences in HbO between my implementation and the nirs-toolbox but I had attributed them to how distances between optodes were calculated. Could you check if the distances that Homer uses are the same as ours? I believe Homer doesn't use 3D coordinates and uses a 2d projection instead. But I would have guessed this would be a 10% difference, not a 500% change. The other issue I recall battling with was the extinction coefficients (#6975 (comment)). At the time I wrote that code we didn't have permission to copy the Homer values so I extracted them myself from the primary source (https://omlc.org/spectra/hemoglobin/summary.html) (but now I think we can include the Homer version of the file? That would be better. Could be a nice outcome of this PR/issue). I think the values I had contained one less decimal point of accuracy. But again I would expect this to be a small effect, not a 5x difference. Can you check that the extinction coefficient we use is the same as Homer? While googling I noticed some differences between Homer and the original reference, but this was already raised by @HanBnrd recently and should be fixed (https://groups.google.com/g/homer-fnirs/c/nRmfgtwQitA, BUNPC/Homer3#6). Johann did you ever compare/validate the output of the MNE BLL to Homer? I think the most likely candidate for this 5x difference is the scaling factor used to convert extinction coefficient to absorption coefficient (its 2.3, so maybe I have applied it twice somewhere and causes a ~5x error?). Anyway I will take a look at this in more detail with you next week. Theres only a few factors in the equation so it shouldn't take me too long to get to the bottom of it. I haven't used Homer before, so could you provide the basic script to convert the raw data to HbO? Then I can add homer to In the meantime the following info may also be helpful (and save me time clicking when back at my PC)... Other info: |
|
Ok curiosity got the better of me and I checked our implementation in master against the latest nirs-toolbox. They seem as similar as before (see https://github.com/rob-luke/mne-fnirs-demo/blob/master/BeerLambert_Validation.ipynb). So I dont think this is a bug that has been recently introduced. I will dig in to the extinction coefficients and distances next week if you can attach a basic homer script. |
|
I just had a look at the extinction coefficients in MNE's However please note that by default in the fixed version of Homer as well as in the old versions between 650 and 900 nm (cf. issue and discussion) Homer uses the extinction coefficient dataset from Wray et al. 1988, while MNE uses Gratzer and Kollias compiled by Scott Prahl. Unfortunately @rob-luke I didn't try to replicate Homer's results with MNE but only with NIRSimple. As I could not use MNE's loaders for our lab's fNIRS devices I created this NIRSimple Python package to apply the modified Beer-Lamber law on any kind of data as long as loaded as a numpy array. When creating it, I spent some time recompiling extinction coefficients from different original sources in CSV files here (references are in the script file) that we could eventually use for MNE if you want? I will try to look more in the detail of the MNE code to see if I can find something. @larsoner could you please share the scripts you used with both MNE and Homer to compare the results (with wavelengths, source-detector distances, extinction coefficient dataset used with Homer, ...)? |
|
MNE was very simple: HOMER I have never personally used, those data came from a collaborator. I'll see if they would be willing to write some lines of MATLAB that should be equivalent, but don't wait for me to do it if it's obvious already to you! EDIT: Changed |
|
This is the HOMER code that was used (untested by me): This is after making the .nirs file with the NIRx conversion script (I assume this is standard/available somewhere else, too?). EDIT: This homer code is incomplete, see below for self-contained code. |
|
Thanks to Johann we have established that the coefficents are correct. And I checked that our code produces results that (approximately, not 5x difference) match the nirs-toolbox beer lambert output (nirs-toolbox is written by Ted Huppert who also wrote Homer [1]). So no alarm bells here. Honestly, I am quite pleased to see how close the two waveforms are!! It means our data reading, channel ordering, optical density, tddr, filtering, BLL, are all working well 😄!! Scaling issues we can fix, its the non linear processing I was worried about. The code you are replicating has quite some preprocessing applied before the Beer Lambert law (BLL). Perhaps it would be best to test the exact same input to the BLL function? Otherwise it could be one of the other steps in the Homer script that is scaling the data? Anyway I dont think the (untested) script above is what produced your results, see details below. I used the provided conversion script to convert our test data to .nirs... >> HomerOfflineConverter('C:\Users\xxxx\mne_data\MNE-testing-data\NIRx\nirx_15_0_recording')
Alert: rdir Matlab function not available. Trying to load single dataset instead.
ProbeInfo file is using the ICBM-152 head model
Dataset converted: C:\Users\xxxxx\mne_data\MNE-testing-data\NIRx\nirx_15_0_recording\NIRS-2019-10-27_003.wl1This successfully produced a .nirs file. I then ran... cd Homer2
setpaths
ADDED search paths for worspace C:\Users\xxxx\Documents\Repositories\homer2
Found fluence files in ./PACKAGES/AtlasViewerGUI/Data/Colin/fw/
load('./PACKAGES/AtlasViewerGUI/Data/Colin/fw/fluenceProf1.mat')
Saving 2 wavelength simulation in ./PACKAGES/AtlasViewerGUI/Data/Colin/fw/fluenceProf1.mat
...... (removed to shorten)
All required toolboxes are installed.
*** For instructions to perform basic tests of Homer2_UI and AtlasViewerGUI, open the PDF file C:\Users\xxxx\Documents\Repositories\homer2/PACKAGES/Test/Testing_procedure.pdf ***
Which seems fine. But when I loaded the .nirs file... load('C:\Users\xxxxxx\mne_data\MNE-testing-data\NIRx\nirx_15_0_recording\NIRS-2019-10-27_003.nirs', '-mat')
>> whos
Name Size Bytes Class Attributes
SD 1x1 3796 struct
aux 92x1 736 double
d 92x20 14720 double
s 92x2 1472 double
t 92x1 736 double It does not contain the variable [1] Huppert, T.J., Diamond, S.G., Franceschini, M.A. and Boas, D.A. (2009). “HomER: a review of time-series analysis methods for near-infrared spectroscopy of the brain.” Appl Opt 48(10): D280-98. |
|
Thanks @HanBnrd for checking the coefficents so quickly!
What type of device do you have? If its a commercially available one is there any chance we can write a loader for it? This might reduce your workload in the long run (e.g. we have both written implementations of cbsi (NIRSimple, MNE)).
This would be great. I am a bit busy in the coming weeks, but after that it might be a nice project if we have your permission to copy code/data from your repositories? |
I think this line will use the Wray et al. 1988 table while
We use Artinis devices and I wanted to help on the loaders when I saw this MNE issue but didn't want to commit to it yet as I cannot access our lab's devices easily because COVID-related restrictions... I'm thinking it could be interesting to have the option to do the preprocessing without channel locations (function
Sure, I'm happy to help if needed. |
Okay in my code with my data I did the reshaping gymnastics to get the HOMER/MATLAB input to the Python version at least:
Agreed that the scripts above are not complete. I'll see if I can figure out how procInput etc. were made next, though I'm not sure we'll need them since Python and HOMER/MATLAB |
Ok this isn't good to hear 😢, so we may have a scaling difference relative to Homer that need to be investigated.
If we can get a homer script that does the most simple processing of... reading raw data, convert to optical density, then beer Lambert law... then I can apply the exact same processing to our tests data and start to dig in to where the differences arise. At the same time I could dig in to the BLL literature again and document our code better. I am writing grants now, but I can explore this once submitted. |
|
Good news! There was just some bug at the HOMER end where distances were accidentally changed during some GUI interactions. Now when reproducing directly with a HOMER script: The Python code above (when used with @rob-luke okay just to merge this as a tiny cleanup? :) |
|
Absolutely fantastic! This is great @larsoner. It will also help to have this small script to show Homer users how easy it is to switch to MNE. Thanks for taking the MATLAB hit for all of us 😃 |
|
@rob-luke do you want to push a commit with some comments about the equations / formulas? Since you had to dig them up recently, maybe now is a good time. |
|
You solved it before I dug out the journal articles again. But I think I'll follow this up in a few weeks and refactor the code to split up the channel wrangling and use a private function just for the BLL. This would allow interested researchers to change things like channel distance and understand the effect on the output. I'll also add the different absorption options from the csv's above. And possibly an age dependent ppf value. But I won't change the public interface, will be all backend changes. And I'll hit the papers then and add comments at the same time. Thanks again for validating this against Homer. I'm very pleased it worked out. |
|
No what’s new entry ? |
|
No, this ended up just doing |
* upstream/master: (66 commits) MRG, ENH: Add infant template downloader (mne-tools#8738) ENH: add reader for NeuroElectrics .nedf files (mne-tools#8734) DOC: improve glossary entry about fiducials (mne-tools#8763) MRG, ENH: Add Report.add_custom_css (mne-tools#8762) BUG, DOC: read_raw_egi didn't support pathlib.Path; update read_raw() docstring (mne-tools#8759) Add "dbs" as new channel type (mne-tools#8739) MRG, VIZ: Fix title position in plot_sensors (mne-tools#8752) MRG: Support for non-FIFF files in Report.parse_folder (mne-tools#8744) MRG, VIZ, FIX: sEEG picking in _prepare_topomap_plot() (mne-tools#8736) DOC: don't use single letter variable name in _compute_forward (mne-tools#8727) WIP: Fix search [skip github] [skip azp] (mne-tools#8742) WIP: Compare Beer-lambert to HOMER (mne-tools#8711) MRG: bump spyder version (mne-tools#8020) FIX anon with IO round trip (mne-tools#8731) fix set_bipolar_reference for Epochs (mne-tools#8728) WIP: Add width argument, reduce default (mne-tools#8725) ENH: Add toggle-all button to Report (mne-tools#8723) fix int/float conversion in nicolet header (mne-tools#8712) MRG, BUG: Fix Report.add_bem_to_section n_jobs != 1 (mne-tools#8713) MRG, DOC: Make "rank" options in docs more accessible (mne-tools#8707) ...
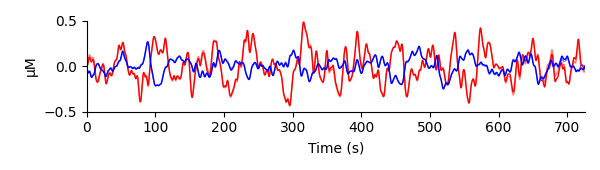
@rob-luke I'm trying to replicate some analysis done in HOMER and I can come pretty close when I use
beer_lambert(raw, ppf=30)(MNE opaque, HOMER translucent):The HOMER analysis used a partial pathlength factor of 6 mm. Any idea why I need to use
ppf=30instead ofppf=6to get a match? Does it have to do with HOMER using a "modified" Beer-Lambert law (MBLL)?Speaking of which, I'm trying to reconcile 4.7 on page 35 of http://www.nmr.mgh.harvard.edu/DOT/resources/homer/HomER_UsersGuide_05-07-28.pdf with our code but I can't. Could you push some commits to add comments and/or point me to some docs that would allow me to put some inline code comments about the equations?
In the meantime this PR is just a cosmetic
(A.T @ B.T).T -> B @ Achange.